Enzyme Engineering
Open Access
ISSN: 2329-6674
ISSN: 2329-6674
Research Article - (2022)Volume 11, Issue 6
Mitochondrial DNA (mtDNA) protein complexes, called mitochondrial-nucleoids (mt-nucleoids), are dynamic and redistribute during mitochondria propagation and gene expression. These proteins involve in mtDNA replication, transcription, maintenance and repair. In this study, we analyzed mt-nucleoid proteins in isolated mt-nucleoids from cotyledons at different developmental ages. We aimed to understand the changes in mt-nucleoids associated protein during cotyledon development. We found that some transcription/translation factors pre-exist in nucleoids of dormant seed mitochondria. High numbers of metabolic proteins co-purified with mtDNA appeared in dormant seeds and developmental cotyledon mitochondria. This report provides some idea about the dynamic of mt-nucleoid proteins along with cotyledon mitochondrial development during seed germination.
Mitochondrial DNA protein complexes; Mitochondrial-nucleoids; Plant mitochondria; Mung bean (Vigna radiata); Mitochondrial development
Mitochondria have their own genome, called mitochondrial DNA (mtDNA). MtDNA is packaged into mtDNA–protein complexes called mitochondrial nucleoids (mt-nucleoids), and the properties of mt-nucleoids change more substantially than the chromosome in the nucleus during development [1]. There are thousands copies of the mitochondrial genome in hundreds of nucleoids in mammalian cells [2,3]. In plants and yeast, more than one nucleoid exists per mitochondrion. These structures are dynamic and redistribute during mitochondria fission /fusion and development [2,4].
Previous studies suggested a multilayer model of mtDNA in mammal cells, with mtDNA replication and transcription occurring in the inner core of the mt-nucleoid. DNA replication- and transcription-related proteins and DNA-packaging proteins, such as mitochondrial transcription factor A (TFAM), single-stranded DNA binding protein (mtSSB), DNA polymerase γ, mtRNA polymerase, and Twinkle, are located in the inner core region [5]. RNA processing and translation occur in the peripheral region that surrounds the core region. The peripheral region is abundant in metabolic proteins, such as heat shock protein 60 (Hsp60), prohibitin, and ATPase family AAA domain-containing protein 3. These proteins may function in mtDNA maintenance and repair [5,6].
The yeast model of mt-nucleoids suggests a segregation apparatus with the core components consisting of a membrane protein complex: mitochondrial distribution and morphology Mdm10-Mdm12 and maintenance of mitochondrial morphology Mmm1 (Mdm10-Mdm12-Mmm1). When yeast cells are in the respiration repressed condition, mtDNA is packaged by ARS-binding factor (Abf2), and mtDNA transactions are inhibited. Levels of aconitate hydratase, cytoplasmic 1 (ACO1) and isoleucine-plus-valine requiring 5 are increased and substitute for the Abf2 packaged during the cell shift to the respiration condition [2]. It has been well studied that mt-nucleoid proteins involved in nucleoids segregation and regulation of gene expression.
Unlike mt-nucleoids in mammal and yeast cells, the content and structure of mt-nucleoids remain unclear in plants. Previous report confirmed that mt-nucleoids have self-directed DNA synthesis ability in-vitro, and this process was inhibited by the DNA polymerase γ inhibitor. It also demonstrated that isolated mt-nucleoids have their own transcriptional system and may perform RNA synthesis ability in vitro [4]. These results strongly support mt-nucleoids in higher plants functioning in mtDNA packaging and segregation and being important for mtDNA maintenance and mitochondrial-encoded gene expression. Ultra-structural analysis of mung bean seed germination showed that cotyledon mt-nucleoids underwent distinctive changes during the early stages of cotyledon development [4].
Here we analyzed mt-nucleoids isolated from cotyledons of different ages. We aimed to understand the dynamics of mt-nucleoid proteins involved in mtDNA structural complexity, replication activity and mitochondrial gene expression in cotyledon development during seed germination. Our LC MS/MS analysis revealed almost 50 proteins associated with mt-nucleoids which were purified without pre-crossing link treatment. Some of these proteins showed dynamic in mt-nucleoids in cotyledons at different ages during seed germination. Several important factors pre-exist in dormant seed mt-nucleoids. Higher levels of metabolic proteins are found which are expected for protecting DNA structure. The results provide some insights about how mtDNA-associated proteins change with cotyledon development during seed germination, which may provide some idea about the initiation of mitochondria gene expression in dormant seeds, the interaction among mt-nucleoid proteins and mtDNA gene expression during seed germination.
Mitochondrial isolation from mung bean cotyledons of different ages
Mung bean Vigna radiata L. (Wilzed) cv Tainan No. 5 was used in this study. Cotyledons of different ages examined included dry seeds, seeds in 4°C water for 12 hr and seeds immersed in water at 27°C for 12 hr. Seedlings were used on days 1, 2 and 3 after 12 hr of water immersion at 27°C, followed by 12, 36 and 60 hr, respectively, growth in vermiculite at 27°C. Besides dry seeds, all embryos were removed. All cotyledons were harvested from seeds, and seedlings were kept in the dark to isolate different-aged mitochondria. We previously described the preparation of the M-system isolation buffer and removal of plastids/nuclei [7]. Pulsed Field Gel Electrophoresis (PFGE) analysis of mitochondria was conducted as we previously described [4].
Mt-nucleoid isolation from mitochondria of mung bean cotyledon at different ages
Mt-nucleoids were isolated from mitochondria of different-aged mung bean cotyledon as described [4]. Sucrose-gradient purified mitochondria were pretreated with high salt (1.2 M KCl) and suspended at 5 mg protein equivalent per milliliter in isolation buffer [15 mM Tris-HCl, pH 7.6, 1.5 mM EDTA, 0.38 M sucrose, 0.6 mM spermidine, 5.3 mM 2-mercaptoethanol, 0.2 mM Phenylmethylsulfonyl Fluoride (PMSF)] and lysed with 0.5% NP-40 and 0.5% sarkosyl. After centrifuging at 14,000 g for 20 min, the clear supernatant was loaded on 4-layer sucrose gradients (15, 30, 50 and 60% w/v) in gradient buffer (20 mM Tris-HCl, pH 7.6, 1 mM EDTA, 1 mM spermidine, 7 mM 2-mercaptoethanol, 0.4 mM PMSF) and centrifuged at 46,000 g for 1 hr. Nucleoids were recovered from the 15 % to 30% sucrose boundary, diluted with 2 vol of gradient buffer and pelleted at 46,000 g for 1 hr.
LC-MS/MS analysis
The pelleted of different stages mt-nucleoid were washed with 25 mM NH4HCO3 solution for 20 min, and then dried in a vacuum centrifuge. Resuspended the protein 50 μl 25 mM NH4HCO3 solution with 12.5 ng/μl trypsin (Sequencing Grade Modified Trypsin, Promega), and incubated for 12 to 16 hr at 37°C. An 50 μl solution containing 5% (w/v) formic acid and 50% (w/v) acetonitrile was added to the pelleted, agitated for 1 h, and moved into a new tube. Then repeated with solution containing 2.5% (w/v) formic acid and 50% (w/v) acetonitrile for 1 hr to to recover the tryptic peptides.
The tryptic peptides were purified with a PepClean C-18 spin column (Pierce). Pellets of tryptic peptides were dissolved in 10 to 20 μl of 0.1 % (w/v) formic acid for LC-MS/MS analysis. Data were analyzed with an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae database (green plants, 28,773 protein entries). The database search for protein identification was performed as described [8]. All stages of data are from duplicate studies.
Immunoblot analysis
Equal amounts of proteins (5 μg) from different stages of cotyledon mitochondria were separated by 12.5 % SDS-PAGE gel, then transferred to polyvinylidene fluoride membranes (Millipore) and incubated with m anti-SSB and anti-ATPaseα antibodies (GTMA) to detect MtSSB and ATPaseα. Immunoblot analysis was performed with the Amersham ECL Plus-Western Blotting Detection System (GE Healthcare). The film was exposed for 30 sec.
PFGE analysis of changed mt-nucleoid DNA movement at different stages of cotyledon development
To investigate mt-nucleoid structure alterations at different stages of cotyledon development, we used PFGE analysis of purified mt-nucleoids from dry seeds, 4°C-12 hr, 27°C-12 hr, day 1, day 2 and day 3. The mtDNA–protein complex (mitochondria lysed and assayed without protease treatment) separated into two sub-populations: one stayed in the sample loading well was defined as the “well-bound DNA protein complex” (wb-DNA protein complex) and the other that migrated as a smeared zone was defined as “the fast-moving DNA protein complex” (fm-DNA protein complex) [4, 9]. The fm-DNA protein complex was present in mt-nucleoids at all stages, from 27°C-12 hr to day-3 cotyledon mitochondria, and migrated to the area equivalent to a 50 to 300-kb DNA position on PFGE analysis. However, under equal amounts of mitochondria (in terms of protein content), the wb-DNA protein complex was found in samples with 27°C 12-hr immersion in water and day 1, 2, and 3 samples but not easily found in dry seeds or 4°C 2,6 and 12-hr water-immersed cotyledons (Figure 1A).
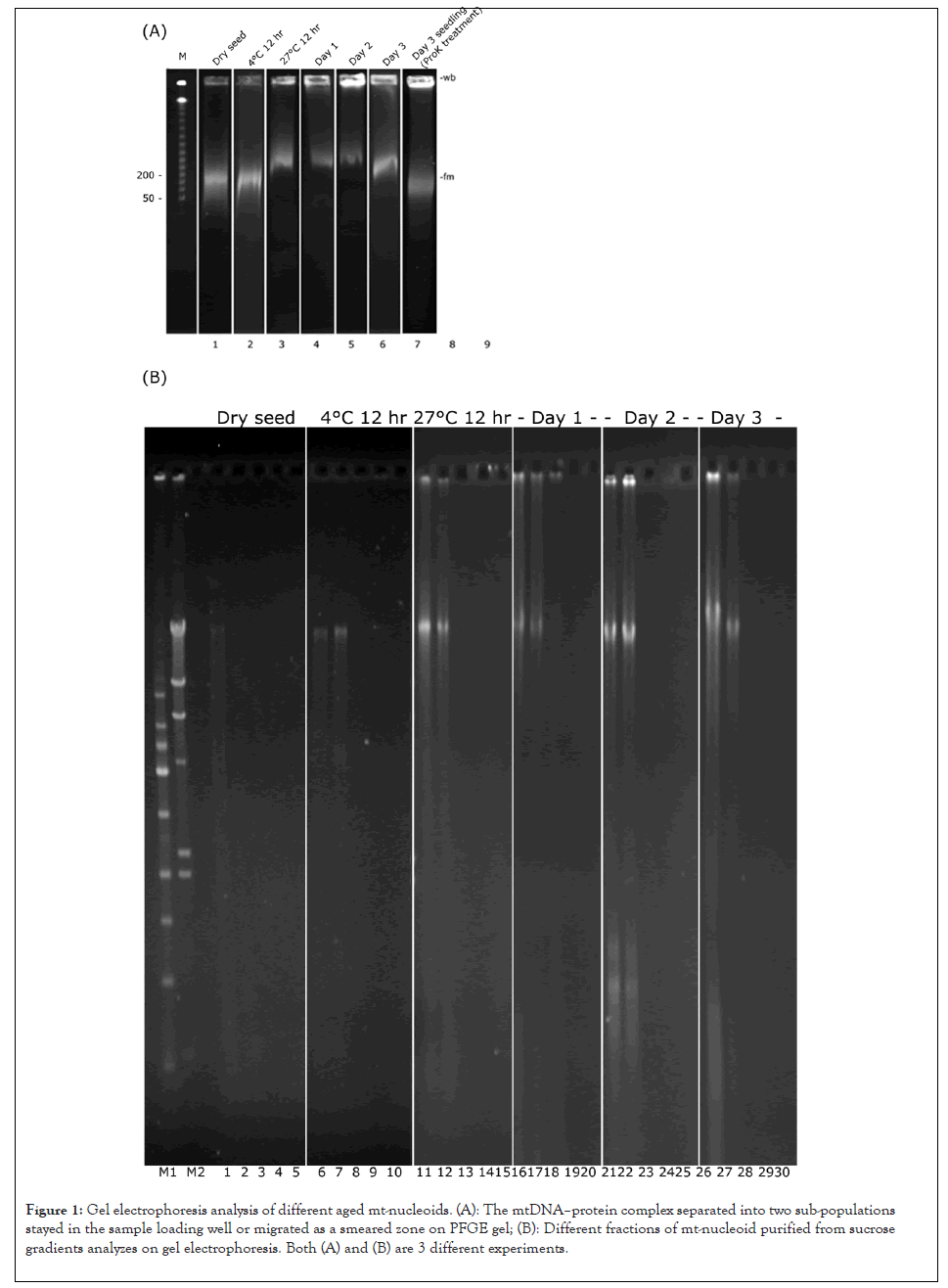
Figure 1: Gel electrophoresis analysis of different aged mt-nucleoids. (A): The mtDNA–protein complex separated into two sub-populations stayed in the sample loading well or migrated as a smeared zone on PFGE gel; (B): Different fractions of mt-nucleoid purified from sucrose gradients analyzes on gel electrophoresis. Both (A) and (B) are 3 different experiments.
To make sure completely gets the pure mt-nucleoid in different-aged cotyledon mitochondria, preliminary testing was carried out by sucrose gradient analysis. Samples were recovered from the supernatant and the 15% to 30%, 30% to 50%, 50% to 60% boundary of the sucrose gradient for each stage of developing cotyledon mitochondria. Day-3 seedling mt-nucleoids were a control. After agarose gel separation on each fraction obtained after sucrose gradient separation, mtDNA appeared only in the supernatant and 15% to 30% boundary collection (Figure 1B). Our previous study also demonstrated that the mt-nucleoids collected from 15% to 30% sucrose gradient had a chromatin-like structure and associated with a membrane component. These mt-nucleoids showed DNA replication and RNA transcription ability [4]. In this study, mt-nucleoids collected from 15 % to 30 % sucrose gradient underwent LC MS/MS analysis.
Mt-nucleoid proteins related to transcription and translation are present in different-aged cotyledon mitochondria during seed germination
Purified mt-nucleoid samples collected from the 15% to 30% boundary of the sucrose gradient were dissolved in tris-buffer for LC-MS/MS analysis. Almost 50 proteins identified were repeatedly associated with mt-nucleoids (Table S1). We classified these proteins according to their potential functions and attempted to understand the possible function of these proteins involved in different-aged mt-nucleoids during seed germination.
60S ribosomal protein appeared in dry seed mt-nucleoids. DEAD-box ATP-dependent RNA helicase and mediator of RNA polymerase II transcription subunits appeared in the stages of dry seeds. DEAD-box ATP-dependent RNA helicase and mediator of RNA polymerase II transcription subunits were also found in 4°C 12-hr samples. They all disappeared in later cotyledon developing stages. These proteins are associated with transcription factors or editing at early stages before seed germination, these important factors may pre-exist in dry seeds and bind with mtDNA during mt-nucleoid purification (Table 1). With seeds at 27°C 12-hr and cotyledon mitochondria executing their function, these transcription factors no longer bind to mtDNA. However, we found the DNA-directed RNA polymerase subunit of transcription in mt-nucleoids of 27°C 12-hr cotyledon mitochondria, so transcription activity is in progress in this stage and new transcription factors can be produced during mitochondria/nucleus functioning.
| Protein | Accession No. | Dry seed | 4°C 12 hr | 27°C 12 hr | Day 1 | Day 2 | Day 3 | Day 3 seedling |
|---|---|---|---|---|---|---|---|---|
| Scorea/match hitsb | ||||||||
| 60S ribosomal protein L18-3 | Q940B0 | 59/2 | - | - | - | - | - | - |
| DEAD-box ATP-dependent RNA helicase 34 | Q9C8J1 | 58/2 | 54/1 | - | - | - | - | - |
| DEAD-box ATP-dependent RNA helicase 37 | Q84W89 | 50/2 | - | - | - | - | - | - |
| Elongation factor Tu | Q9ZT91 | - | - | 158/5 | 81/2 | 91/2 | 61/2 | 132/3 |
| DNA-directed RNA polymerase subunit beta~ | Q85C71 | - | - | 54/6 | - | - | - | - |
| Mediator of RNA polymerase II transcription subunit 37a | Q9LKR3 | 131/2 | 119/5 | - | - | - | - | 132/5 |
| Lon protease homolog | A2YQ56 | - | - | - | 222/6 | - | 47/2 | - |
| Single-stranded DNA-binding protein | Q84J78 | - | - | - | - | - | - | 64/1 |
Note: Data were analyzed with an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae database (green plants, 28,773 protein entries). Matched proteins were filtered with a significant threshold (P value) of <0.009 (as the methods of Lo et al., 2011).
a. The protein score from a MASCOT MS/MS search derived from the matched peptide ion scores.
b. The match hits from a MASCOT MS/MS search derived from the amount of matched peptides sequence.
Table1: Nucleoproteins related to transcription and translation present in mung bean cotyledon mitochondria with progressive mitochondrial.
Other factors associated to mtDNA that could be found in 27°C 12-hr to day-3 data included elongation factor Tu and Lon protease homolog protein, which suggests that these proteins may bind with mtDNA directly or indirectly in active states of cotyledon mitochondria.
Mt-nucleoid-associating membrane proteins
It is known that mtDNA anchors to the inner mitochondrial membrane by nucleoid proteins. In our results, some membrane-binding proteins, such as ATPaseα, ATPaseβ, and Adenine Nucleotide Transporter 1 (ANT1), appeared in mt-nucleoids at all developing stages (Table 2). ANT and ATPase alpha were not found in day 3 cotyledon mt-nucleoids. MtDNA may directly or indirectly bind with inner membrane proteins. Outer-membrane protein Porin was also found in mt-nucleoids at all stages as previously found [4].
| Protein | Accession No. | Dry seed | 4°C 12 hr | 27°C 12 hr | Day 1 | Day 2 | Day 3 | Day 3 seedling |
|---|---|---|---|---|---|---|---|---|
| Scorea/match hitsb | ||||||||
| ADP, ATP carrier protein (ANT) | P25083 | - | 105/9 | 481/27 | 96/9 | 141/6 | - | 245/17 |
| ATP synthase subunit alpha | P24459 | 682/48 | 731/50 | 919/76 | 388/28 | 664/52 | - | 1172/60 |
| ATP synthase subunit beta | P19023 | 572/27 | 425/24 | 1503/80 | 392/27 | 420/27 | 416/25 | 643/26 |
| ATP synthase subunit gamma | Q96250 | - | - | 216/9 | 83/2 | 84/2 | 107/2 | 99/2 |
| Probable aquaporin PIP-type 7a | P25794 | 62/3 | 87/3 | 153/16 | 91/10 | 65/4 | - | 48/3 |
| Probable aquaporin TIP3-2 | O22588 | 48/2 | 177/8 | 86/5 | 73/3 | 94/3 | 75/2 | |
| Mitochondrial outer membrane protein porin 1 | Q9SRH5 | 161/7 | 116/6 | 332/13 | 100/2 | 77/8 | 80/4 | 135/5 |
| Outer plastidial membrane protein porin | P42054 | 63/5 | 84/3 | 162/23 | 70/2 | 50/2 | 82/4 | 57/3 |
Note: Data were analyzed with an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae database (green plants, 28,773 protein entries). Matched proteins were filtered with a significant threshold (P value) of <0.009 (as the methods of Lo et al., 2011).
a. The protein score from a MASCOT MS/MS search derived from the matched peptide ion scores.
b. The match hits from a MASCOT MS/MS search derived from the amount of matched peptides sequence.
Table 2: Membrane proteins present in mung bean cotyledon mitochondria mt-nucleoids with progressive mitochondrial development during seed germination.
Metabolic proteins related to respiration appear in mt-nucleoids from dry seeds to day-3 cotyledons
Numerous metabolic proteins appeared in mt-nucleoids from dry seeds to day-3 cotyledons (Table 3). Most of these metabolic proteins are related to respiration: complex I protein NADH dehydrogenase, complex II protein succinate dehydrogenase, complex III protein cytochrome c1, and complex IV protein cytochrome c oxidase. We also found glycolysis intermediate proteins: dihydrolipoyl dehydrogenase, superoxide dismutase, dihydrolipoyllysine-residue acetyltransferase component 2, serine hydroxymethyltransferase 1, and citric acid cycle intermediate protein: malate dehydrogenase, aconitate hydratase 2, formate dehydrogenase 1, isocitrate dehydrogenase, succinyl-coa ligase (probable), pyruvate dehydrogenase E1 component subunit beta and succinate-semialdehyde dehydrogenase. We found no evidence that those metabolic-associated nucleoid proteins are directly related to mtDNA maintenance.
| Protein | Accession No. | Dry seed | 4°C 12 hr | 27°C 12 hr | Day 1 | Day 2 | Day 3 | Day 3 seedling |
|---|---|---|---|---|---|---|---|---|
| Scorea/match hitsb | ||||||||
| Cytochrome c oxidase subunit 1 | P07506 | - | - | 179/2 | 93/2 | - | - | - |
| Cytochrome c oxidase subunit 2 | P32646 | 200/9 | 171/9 | 119/11 | 96/8 | - | 56/6 | 269/16 |
| Cytochrome c1-1, heme protein | P25076 | 40/4 | 67/6 | 65/5 | 41/5 | 96/4 | 54/4 | 63/3 |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9 | Q9SK66 | 87/2 | - | 45/3 | 82/2 | - | - | 57/2 |
| NADH dehydrogenase (ubiquinone) flavoprotein 1 | Q9FNN5 | 142/6 | 74/2 | 78/8 | 127/6 | 137/4 | 154/4 | 60/4 |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 1 | Q9FGI6 | 115/4 | - | 92/6 | 108/5 | 79/2 | - | 113/5 |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 2 | P93306 | 68/5 | 79/4 | 166/8 | 110/6 | 123/6 | 121/5 | 68/4 |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 3 | Q33994 | 80/6 | - | - | - | 64/7 | 44/2 | - |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 8 | P80269 | 75/2 | 96/3 | - | - | - | - | 145/2 |
| Dihydrolipoyl dehydrogenase (Fragment) | P80503 | 125/2 | 110/2 | 118/2 | 121/3 | 141/2 | 146/3 | 129/2 |
| Dihydrolipoyl dehydrogenase | P31023 | - | - | 130/10 | 92/5 | 61/4 | 99/5 | 173/6 |
| Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex | Q8RWN9 | - | - | 82/8 | 99/4 | 107/4 | 89/3 | 59/4 |
| Malate dehydrogenase | P17783 | 163/7 | 179/7 | 268/16 | 182/16 | 230/10 | 304/19 | 329/14 |
| Aconitate hydratase 2 | Q9SIB9 | 90/2 | 37/2 | - | - | 68/2 | 123/3 | 330/16 |
| Formate dehydrogenase 1 | Q9SXP2 | 127/2 | 83/2 | 144/4 | 138/5 | 93/5 | - | 130/2 |
| Isocitrate dehydrogenase (NAD) catalytic subunit 5 | Q945K7 | - | - | 61/4 | - | 65/2 | - | 77/2 |
| Isocitrate dehydrogenase (NAD) regulatory subunit 1 | Q8LFC0 | 125/4 | 58/2 | 408/21 | 161/6 | 47/5 | 88/6 | 58/3 |
| Isocitrate dehydrogenase (NAD) regulatory subunit 3 | O81796 | - | - | 60/9 | 71/4 | - | 116/4 | - |
| Probable succinyl-CoA ligase (ADP-forming) subunit alpha | Q6ZL94 | - | - | 82/2 | 55/2 | - | 56/4 | 54/2 |
| Pyruvate dehydrogenase E1 component subunit beta | P52904 | 69/2 | 56/2 | 84/4 | 67/2 | 84/4 | 52/2 | 255/8 |
| Serine hydroxymethyltransferase 1 | P49357 | - | 52/2 | 104/11 | 97/5 | 76/6 | 51/4 | 115/4 |
| Succinate dehydrogenase (ubiquinone) flavoprotein subunit 1 | O82663 | 184/6 | 122/6 | 178/14 | 239/12 | 225/10 | 127/9 | 48/6 |
| Succinate-semialdehyde dehydrogenase | B9F3B6 | - | - | 74/2 | 45/5 | - | 77/2 | - |
| Superoxide dismutase (Mn) | P35017 | - | 40/2 | 77/8 | 140/3 | - | 73/2 | 83/2 |
Note: Data were analyzed with an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae database (green plants, 28,773 protein entries). Matched proteins were filtered with a significant threshold (P value) of <0.009 (as the methods of Lo et al., 2011).
a. The protein score from a MASCOT MS/MS search derived from the matched peptide ion scores.
b. The match hits from a MASCOT MS/MS search derived from the amount of matched peptides sequence.
Table 3: Metabolic proteins related to respiration present in mung bean cotyledon mitochondria mt-nucleoids with progressive mitochondrial development during seed germination.
Other proteins found in cotyledon mt-nucleoids functioning in protein quality control reported in mammal cells and yeast studies (Table 4). Chaperonin 60 (CPN60) belonging to Hsp60 appeared in 4°C 12 hr to day 3 cotyledon mt-nucleoids. CPN60 is involved in mitochondrial protein homeostasis functioning in protein refolding [10]. Hsp70 was also found in human and yeast mt-nucleoids in previous studies [10,11]. It was found in mt-nucleoids during the whole period of cotyledon mitochondria development we investigated.
| Protein | Accession No. | Dry seed | 4°C 12 hr | 27°C 12 hr | Day 1 | Day 2 | Day 3 | Day 3 seedling |
|---|---|---|---|---|---|---|---|---|
| Scorea/match hitsb | ||||||||
| Delta-1-pyrroline-5-carboxylate dehydrogenase 12A1 | Q8VZC3 | - | - | 66/9 | - | - | - | 51/2 |
| Aldehyde dehydrogenase family 2 member B4 | Q9SU63 | 40/3 | 55/4 | 44/6 | 79/4 | 75/8 | 79/7 | 44/3 |
| Chaperonin CPN60 | P35480 | 150/11 | 161/7 | 426/36 | - | 111/14 | - | 306/18 |
| Chaperonin CPN60-1 | P29185 | 250/18 | 196/11 | 797/54 | 394/24 | 229/18 | 154/20 | 342/25 |
| Chaperonin CPN60-2 | Q05046 | 131/15 | - | 773/41 | 407/26 | 291/20 | 235/19 | 374/24 |
| Heat shock 70 kDa protein | Q01899 | 218/13 | 96/7 | 552/40 | 381/25 | 156/12 | 107/10 | 421/10 |
| Catalase isozyme 2 | P12365 | - | 37/3 | - | - | - | 53/4 | - |
| Catalase-4 | O48561 | 51/2 | 52/3 | - | 48/2 | - | 59/3 | 159/9 |
| Nucleoside diphosphate kinase III | O49203 | - | - | - | - | 53/3 | 69/2 | - |
| Methylcrotonoyl-CoA carboxylase subunit alpha | Q42777 | - | 43/2 | -- | 63/2 | 69/2 | 38/3 | 73/3 |
| Isovaleryl-CoA dehydrogenase 2 | Q9FS87 | - | - | 65/3 | 65/3 | 46/1 | 84//4 | - |
Note: Data were analyzed with an in-house Mascot search program (v. 2.2.06; Matrix Science) against the UniProtKB/Swiss-Prot Viridiplantae database (green plants, 28,773 protein entries). Matched proteins were filtered with a significant threshold (P value) of <0.009 (as the methods of Lo et al., 2011).
a. The protein score from a MASCOT MS/MS search derived from the matched peptide ion scores.
b. The match hits from a MASCOT MS/MS search derived from the amount of matched peptides sequence.
Table 4: Metabolic proteins with functions in protein quality control in mung bean cotyledon mitochondria mt-nucleoids with progressive mitochondrial development during seed germination.
Catalase is peroxisomal enzyme and functions in Reactive Oxygen Species (ROS) homeostasis. It is found in peroxisomes and also in mitochondria in yeast, maize, Arabidopsis and animal cells [12,13]. We found the homolog in dry seeds, 4°C 12 hr, day 1 and 3 cotyledon mt-nucleoids (Table 4), which indicates a high level of ROS involved during these stages.
Our previous report suggested that the structural complexity of mtDNA and its replication activity change with cotyledon development during seed germination [9]. This change may reflect mtDNA replication, gene expression, and mtDNA repair during cotyledon development. Our results indicate that changes in mt-nucleoid proteins do exist in different-aged cotyledons during seed germination by LC MS/MS analysis. Mt-nucleoids, MtDNA-protein complex, shows dynamic alteration along cotyledon development also exhibits in gel analysis (Figures 1A and 1B) which indicates that mtDNA and its associated proteins may change their interaction and result in composition/conformation alteration. Some transcription and translation related proteins also found in mt-nucleids in dormant and pre-germination (4°C 12-hr) seeds.
Single-stranded DNA binding protein (mtSSB) is a marker protein in mt-nucleoid studies and is involved in mitochondrial DNA replication [14,15]. Mung bean cotyledon mitochondria showed high DNA replication activity in samples with 27°C 12-hr immersion in water to day 2 after seed germination [9], but we did not find mtSSB on LC MS/MS analysis of isolated mt-nucleoids from any age cotyledon even though it was found in day-3 seedling mt-nucleoids by proteomic analysis (Table 1). MtSSB could be detected in cotyledon mitochondria by western blot analysis (Figure S1). We suspect that the amount of mtSSB protein may be too low to be detected due to our cotyledon mt-nucleoid purification process without pre-crosslink treatment. We also could not find DNA polymerase γ in our study, which was shown in our previous work [4,9], the lack of DNA polymerase γ in mt-nucleoid proteomic data may also be due to its too-low content in purified cotyledon mitochondria.
According to our previous results, mt-nucleoids from mung bean day-3 seedlings had transcription ability in vitro. TFAM (mitochondrial transcription factor A) is another essential protein found in mt-nucleoids that directly binds with mtDNA in mammalian cells and is involved in transcription [6,16], but it has no homolog in plant cells. In Arabidopsis, as compared with TFAM, the T7 bacteriophage RNA polymerase is involved in all steps of transcription, including promoter recognition, initiation, and elongation, as a single-polypeptide enzyme in mitochondria [17]. T7 bacteriophage RNA polymerase (DNA-directed RNA polymerase) was found in mt-nucleoids of 27°C 12-hr cotyledon mitochondria by our proteomic analysis, which suggests that mt-nucleoid initiates its transcription when seeds immersed in 27°C 12-hr water and starts its DNA gene expression (Table 1).
Some nucleoproteins related to transcription and translation were found: the DEAD-box RNA helicase which participate in RNA metabolism and use ATP to bind or remodel RNA for involvement in mechanisms such as RNA splicing, transcription initiation, ribosome assembly, or nuclear export [18]. Recent plant genome and proteome analyses suggest that DEAD-box RNA helicases in mitochondria are crucial for regulating gene expression and RNA metabolism. Several studies have demonstrated that mitochondria-localized DEAD-box RNA helicases in rice and Arabidopsis can regulate the expression of stress-induced genes and seed germination [19]. DEAD-box ATP-dependent RNA helicase appeared in dry seeds and 4°C 12-hr samples in our results, which suggests that the RNA helicase homologs of mung bean cotyledons stay in dormancy seeds and prepare for initiating mitochondrial gene expression before seed germination.
We suggest that when dormant seeds start to germination, cotyledon mitochondrial genes will turn on immediately by using the pre-existing transcription/translation factors and ribosomal protein. After first step of mt gene expression completed, the pre-existed ribosomes and DEAD-box ATP-dependent RNA helicase etc. are no longer associated with mt-nucleoids. Instead, the DNA-directed RNA polymerase subunit of transcription turns on in 27°C 12-hr cotyledon mt-nucleoids and transcription activity is in progress functioning for seeds germination (Table 1).
We also found that the elongation factor Tu for translation bound with mtDNA directly or indirectly in active states (from 27°C 12-hr to day 3) of cotyledon mitochondria (Table 1). The transcription-related protein ATP-dependent protease Lon is appeared in day 1 and day 3 but not day 2 cotyledon mt-nucleoids. The reason for Lon missing in day 2 mt-nucleoid is not clear. Previous study shown that Lon was located in the central region in mt-mucleoid in mammalian cells and was involved in TFAM degradation and mtDNA quality control [20]. ANT1 and voltage-dependent anion channel may form complexes and generate contact sites between the mitochondrial inner and outer membranes (Table 2) [4,21]. Above results imply that mt-nucleoids anchor directly or indirectly with the membrane contact site.
Mt-nucleoids are known to bind to the inner mitochondrial membrane. The outer membrane porin is found in mt-nucleoids as previous report [4]. The appearance of porin in the LC MS/MS data suggested that mt-nucleoids may bind directly or indirectly on the membrane contact site (Table 2). Many metabolic proteins, including a large amount of respiration-related proteins, appear in cotyledon mt-nucleoids from dry seeds to day-3 seedlings. Some appear in day-1 to -3 cotyledon and day 3 seedlings (Table 3). Recent research suggested that respiration-related metabolic proteins found in yeast and human mt-nucleoids may be involved in organizing and protecting mtDNA against oxidative damage during respiration [2,5]. We expect that higher numbers of metabolic proteins appeared in mt-nucleoids along cotyledon development may also play the dual functions in protecting the complex DNA structure when respiration is active in mitochondria. Same phenomenon is also found in mammalian and yeast. Recent studies suggest that more metabolic proteins associate with mt-nucleoids when mitochondria have higher physiology activity, and these proteins may be involved in organization and protection of mtDNA against oxidative damage during respiratory growth. The studies suggested that these bifunctional metabolic proteins participate in both mitochondrial metabolism and mtDNA maintenance [3]. For example, the ACO1 gene encoding the aconitase 1 deletion in yeast induced the loss of mtDNA. Thus aconitase may have a direct role in physical protection of mtDNA [22,23]. Aconitase 1 found in dry seeds, 27°C 12-hr, day 2 and day 3 cotyledon mt-nucleoids suggests that metabolic proteins such as aconitase 1 in mt-nucleoids may be bifunctional in mtDNA protection.
Proteins that function in protein quality control are easily found in mammalian and yeast mt-nucleoids. Recent studies suggest that Hsp60 functions in mitochondrial protein import and interacts with the mtDNA ori sequence and is required for nucleoid division [17] (Kuhn et al. 2007). We found CPN60 and Hsp70 in cotyledon mt-nucleoids at all developing stages (Table 4). However, whether these quality-control proteins play a specific role in nucleoid organization or only in protein import and protein-quality control is unclear [10,11].
Similar to most studies, our proteome data revealed many proteins not predicted to localize to mitochondria and not characterized with respect to nucleoid function. For example, catalase is a peroxisomal enzyme and functions in ROS homeostasis. This protein is also found in peroxisomes and also in mitochondria in yeast, maize, Arabidopsis and animal cells [12]. It was also found in our results (Table 4). The possibility of peroxisome contamination during mt-nucleoid purification should not be ruled out [12].
Some yeast studies suggested that mtDNA maintenance and segregation depend on the actin cytoskeleton [2]. Reyes et al. found that ꞵ-actin belongs to the protease-resistant population and co-fractionates in an iodixanol gradient with human mtDNA, so actin could play a role in mtDNA maintenance [24]. Our previous LC MS/MS study demonstrated that actin is bound to the mtDNA–nucleoprotein complex in mung bean seedlings [8]. However, actin was not found in any stage of cotyledon mt-nucleoids in this study. Actin and other cytoskeletal associated proteins were found in mt-nucleoids in HeLa cells with cross-linking and glycerol gradients in the rapidly sedimenting fraction. [6,25]. Actin may link with the mt-nucleoid complex, but the binding mechanism may be different from that in other nucleoid proteins. Most likely, actin is not found in isolated cotyledon mt-nucleoids because of the long process of cotyledon mt-nucleoid purification without a pre-cross-linking process and special actin protection procedure.
Our results demonstrate that the plant mt-nucleoid proteome is dynamic (Figure 2). Some important factors are stored and pre-exist in nucleoids of dormant seed mitochondria. These stored mt-nucleoid proteins may play an important role for preparing the initiation of mt-DNA gene expression before seed germination.
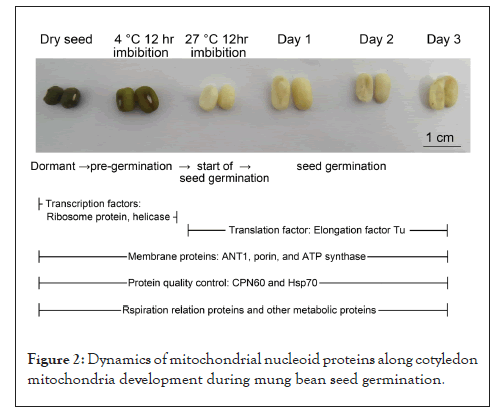
Figure 2: Dynamics of mitochondrial nucleoid proteins along cotyledon mitochondria development during mung bean seed germination.
The essential initiation of gene expression accomplished the pre-existed factors found in dry seeds; mt-nucleoids are released from nucleoids. Then the DNA-directed RNA polymerase turns on followed by mtDNA gene expression for seeds germination. This investigation provides some information on dynamic of mt-nucleoids proteins along cotyledon development. These proteins bind directly and/or indirectly to mtDNA and play a role in mtDNA replication and gene regulation during seed germination. Interestingly, some ribosome protein, helicase and transcription factor are found in dormant seed prepared for initiation of mitochondrial gene expression before seed germination.
This work was supported by research grants from the Ministry of Science and Technology and from National Taiwan University- Institute of Plant and Microbial Biology, Academia Sinica, PhD Program, Republic of China. We thank Dr Tuan-Nan Wen and Proteomics Core Lab in Institute of Plant and Microbial Biology for their help with LC-MS/MS analysis.
No any interests that are directly or indirectly related to the work submitted for publication.
N.C. performed most of the experimental work including isolation of the cotyledon mitochondria, mt-nucleoids isolation, Immunoblot analysis and LC-MS/MS analysis. Y.-S.L. performed Immunoblot analysis, LC-MS/MS analysis and helped in the isolation of cotyledon mitochondria; H.D. and N.-S.L. joined the discussions and provided valuable advice on this research project; N.C. wrote the paper; N.C. composed the figures and tables.
This work was supported by research grants from the Ministry of Science and Technology and from National Taiwan University – Institute of Plant and Microbial Biology, Academia Sinica, PhD Program, Republic of China.
Data generated or analyzed during this study are provided in full within the published article.
Citation: Cheng N, Lo Y, Lin N, Dai H (2022) Dynamics of Mitochondrial Nucleoid Proteins Involved in Development of Cotyledon Mitochondria During Mung Bean Seed Germination. Enz Eng. 11:201
Received: 01-Nov-2022, Manuscript No. EEG-22-20708; Editor assigned: 03-Nov-2022, Pre QC No. EEG-22-20708 (PQ); Reviewed: 18-Nov-2022, QC No. EEG-22-20708; Revised: 25-Nov-2022, Manuscript No. EEG-22-20708 (R); Published: 02-Dec-2022 , DOI: 10.35248/2329-6674.22.11.201
Copyright: © 2022 Cheng N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.