Research Article - (2016) Volume 2, Issue 1
Effect of GPD2 and PDC6 Deletion on Isobutanol Titer in Saccharomyces Cerevisiae
*Corresponding Author: Aili Zhang, School of Chemical Engineering and Technology, Hebei University of Technology, Guangrong Road No 8, Hongqiao District, Tianjin, 300130, People’s Republic of China, Tel: 86-22-60200444 Email:
Abstract
Objectives: Isobutanol is regarded as a next-generation biofuel for its higher octane number and higher energy density than ethanol. However, during isobutanol biosynthesis, ethanol and glycerol are major unwanted byproducts. In order to improve isobutanol production in Saccharomyces cerevisiae, we used molecular biology and genetic recombination technologies to eliminate ethanol and glycerol titers.
Methods: In this study, GPD2 and PDC6 were deleted to increase isobutanol production in microaerobic fermentation of Saccharomyces cerevisiae. Engineered strain HZAL–13 (PGK1p–BAT2 gpd2Δ::RYUR) was constructed by overexpressing of BAT2 (which encodes a branched-chain amino-acid aminotransferase) and deleting GPD2 (which encodes glycerol-3-phosphate dehydrogenase). Engineered strain HZAL–14 (PGK1p–BAT2 pdc6Δ::R gpd2Δ::RYUR) was obtained by further deleting PDC6 (which encodes pyruvate decarboxylase) in HZAL– 13 pILV2. Then we tested the fermentation performances of engineered strains and control strain. During microaerobic fermentation, cultures were performed at 30°C in the unbaffled shake flasks kept at constant stirring speed of 100 rev/min with 100 ml medium for 48 hours.
Results: The maximum isobutanol titers of control strain, HZAL–13 pILV2 and HZAL–14 pILV2 were 29.8 mg/l, 162.3 mg/l and 309.3 mg/l, respectively. These results demonstrate that decreasing glycerol formation and ethanol biosynthesis in combination through deletion of PDC6 and GPD2 could increase dramatically the isobutanol titer in S. cerevisiae.
Conclusion: Overexpression of related genes in isobutanol biosynthesis pathway and deletion of key genes that encode glycerol and ethanol biosynthesis is a promising strategy to increase isobutanol titer in Saccharomyces cerevisiae.
Keywords: Saccharomyces cerevisiae; Isobutanol; Ethanol; Glycerol; LDP1; GPD2; PDC6
Introduction
Environmental friendly biofuels gain great interests due to climate change and the need for renewable transportation fuels in recent years [1]. Ethanol has been received more attention as the most widely used biofuel [2]. However, compared to ethanol, higher alcohols have several advantages as next-generation transport fuels. And they are compatible with current infrastructure [3]. Isobutanol exhibits superior physicochemical properties such as higher energy density and lower hygroscopicity than ethanol [4,5]. Furthermore, it has higher octane number than the isomer n-butanol [6,7].
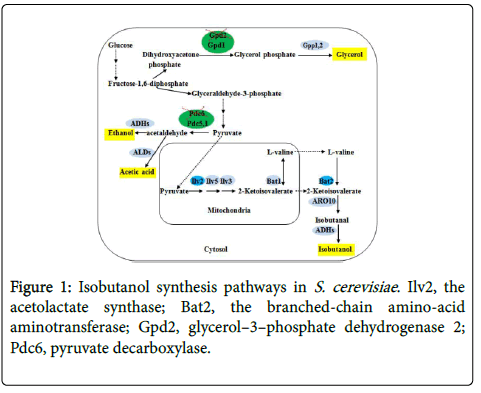
Saccharomyces cerevisiae has high tolerance to isobutanol up to 20 g/l and robustness during fermentation in harsh industrial conditions [8-10]. In addition, S. cerevisiae naturally produces small amount of isobutanol as byproduct from the catabolism of amino acids [11,12]. So S. cerevisiae has been considered as an attractive alternative host strain to produce isobutanol. Initially, glucose is converted to pyruvate via the process of glycolysis. Then pyruvate is converted to 2- ketoisovalerate (KIV) in the mitochondria by acetolactate synthase (Ilv2), acetohydroxyacid reductoisomerase (Ilv5) and dihydroxyacid dehydrates (Ilv3) [12].
After that, KIV is further converted to L–valine by branched-chain amino-acid aminotransferase (Bat1) [10]. Furthermore, L–valine undergoes conversion to KIV by branched-chain amino-acid aminotransferase (Bat2). Finally, KIV converted to isobutanol (Figure 1).
Ilv2 catalyzes the first reaction in the synthesis of KIV in the mitochondria. KIV is the immediate precursor of valine [1]. BAT2 plays an important role in catalyzing the first reaction in the catabolism of valine in the cytosol [13]. PDC6 encodes pyruvate degradation in ethanol biosynthesis and GPD2 encodes glycerol phosphate formation. Carbon source flux mainly flows to ethanol and glycerol biosynthesis in isobutanol fermentation in S. cerevisiae [14]. To further improve isobutanol titer, we constructed engineered strains HZAL–13 (PGK1p–BAT2 gpd2Δ::RYUR) and HZAL–14 (PGK1p– BAT2 pdc6Δ::R gpd2Δ::RYUR). And the fermentation characters of these engineered strains were also investigated. HZAL–13 pILV2 exhibited a 445% increase (162.3 mg/l) in isobutanol production compared with control strain. And the maximum isobutanol titer of HZAL–14 pILV2 showed a 938% increase (309.3 mg/l) than that of control strain.
Materials and Methods
Yeast strains and media
The S. cerevisiae strains used in this study were all isogonics to W303–1A as described in table 1.
| Strains | Complete Genotype | Reference or source |
|---|---|---|
| W303–1A | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100 | Thomas and Rothstein, 1989[15] |
| HZAL–13 | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100 PGK1p-BAT2 gpd2D, RYUR | This study |
| HZAL–14 | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100 PGK1p-BAT2 pdc6Δ:: R gpd2Δ, RYUR | This study |
| W303–1A YEplac181 | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100YEplac181–PGK1p–ILV2 | This study |
| HZAL–13 pILV2 | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100 PGK1p-BAT2 gpd2Δ, RYUR YEplac181–PGK1p–ILV2 | This study |
| HZAL–14 pILV2 | MATa leu2-3, 112 ura3-1 trp1-92 his3-11, 15 ade2-1 can1-100 PGK1p-BAT2 pdc6Δ:: R gpd2Δ::RYUR YEplac181–PGK1p–ILV2 | This study |
Table 1: Strains used in this study.
The strains of S. cerevisiae cells were routinely grown in medium containing 2% peptone (w/v) and 1% yeast extract (w/v) supplemented with 2% glucose (w/v) as carbon source (YPD). Selective media SC [16] minus leucine or uracine were used for selection of transformants containing LEU2 or URA3 selective marker.
Growth conditions and experimental procedures
Incubation conditions were standardized at 30°C and 200 rev/min orbital shaking. Standard techniques were applied as described in Sambrook et al. [17]. For all gene–cloning experiments, PCR products were purified using phenol deproteinization and ethanol precipitation. Restriction and modification enzymes were used according to the manufacturers’ instructions. Yeast transformation was performed by the lithium acetate method. Escherichia coli Top10' was used for subcloning. All yeast strains were maintained at 4°C on YPD plates and prepared monthly from a glycerol stock kept at –75°C.
Fermentation conditions
Microanaerobic cultivations were performed at 30°C in the unbaffled shake flasks kept at constant stirring speed of 100 rev/min with 100 ml medium (selective media SC minus leucine supplemented with 4% glucose (w/v) as carbon source). Initial biomass concentrations were set at OD600nm 0.2 after inoculations. Fermentation experiments were performed in triplicate and one representative experiment was shown.
Plasmids and strains constructions
The plasmids and primers used in this study were described in Table 2 and Table 3, respectively.
| Plasmids | Description | Reference or source |
|---|---|---|
| YEplac181 | AmprLEU2 | Gietz and Sugino [18] |
| YIplac211 | AmprURA3 | Gietz and Sugino [18] |
| pUC18–RYUR | Ampr | Zhang [19] |
| YEplac181–PGK1p–ILV2(pILV2) | AmprLEU2 | This study |
| YIplac211–BAT2p–PGK1p–BAT2 | AmprURA3 | This study |
| pUC18–PDC6P–RYUR–PDC6T | AmprURA3 | This study |
| pUC18–GPD2P–RYUR–GPD2T | AmprURA3 | This study |
Table 2: Plasmids used in this study.
| Primers | Oligonucleotide |
|---|---|
| ILV21to20-U | 5'-GGATCCGTCGACATGATCAGACAATCTACGCT-3' |
| ILV2ORF-D | 5'-GGGCCCCTGCAGCGTTTAGCTGGCTCCTGATG-3' |
| BAT2promoter-U | 5'-GGGCCCGAGCTCCTCTTGTCTACAACACCAGC-3' |
| BAT2promoter-D | 5'-GGGCCCGGATCCTCTAGGGGTGCCAAGGTCAT-3' |
| BAT2ORF-U | 5'-GGGCCCGTCGACATGACCTTGGCACCCCTAGA-3' |
| BAT2ORF-D | 5'-GGGCCCCTGCAGATACCGATAGGCCAGCACTA-3' |
| PGK1 promoter-U | 5'-GGGCCCGGATCCAGGCATTTGCAAGAATTACTC-3' |
| PGK1 promoter-D | 5'-GGGCCCGTCGACTGTTTTATATTTGTTGTAAA AAGTAG-3' |
| CPDC6P-U | 5'-GGGCCCGAATTCATGCAGATCGGCTGTGGCAT-3' |
| CPDC6P-D | 5'-GGGCCCGGATCCGCTCGCGAATCGCACCATAT-3' |
| CPDC6T-U | 5'- CCCGGGCTGCAGAAGCCATTAGTAGTGTACTC-3' |
| CPDC6T-D | 5'-CCCGGGAAGCTTGAGCCAAAGAGATGAGCCAA-3' |
| GPD2P-U | 5'-CCCGGGAAGCTTCAGACGCAGCAGCAAGTAAC-3' |
| GPD2P-D | 5'-CCCGGGCTGCAGGTAGAGAAGAGCTGCTGAAC-3' |
| GPD2T-U | 5'-GGGCCCGGATCCAGGCAGTCTACCAGATAGTCT-3' |
| GPD2T-D | 5'-GGGCCCGAATTCTGACTGGAGAGCCGTCAGTA-3' |
| Relevant restriction sites are underlined. | |
Table 3: Primers used in this study.
Plasmid YA3in this study were all isogon-Iplac211–BAT2p– PGK1p–BAT2 construction
PGK1 promoter–U containing restriction enzyme site for BamHI in front of nucleotides 721 bp to 701 bp upstream of the start codon of PGK1, and PGK1 promoter–D containing restriction enzyme site for SalI in front of nucleotides 1 bp to 26 bp of the complementary strand upstream of the start codon of PGK1, were used to clone parts of the promoter gene of PGK1 by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with SalI and BamHI, and ligated into the SalI and BamHI digestion sites of the integrative plasmid YIplac211, resulting in the plasmid YIplac211-PGK1p.
BAT2 promoter–U containing restriction enzyme site for SacI in front of nucleotides 942 bp to 923 bp upstream of the start codon of BAT2, and BAT2 promoter–D containing restriction enzyme site for BamHI in front of nucleotides 1 bp to 20 bp of the complementary strand upstream of the start codon of BAT2, were used to clone parts of the promoter gene of BAT2 promoter by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with SacI and BamHI, and ligated into the SacI and BamHI digestion sites of the plasmid YIplac211–PGK1p, resulting in the plasmid YIplac211– BAT2p–PGK1p.
BAT2 ORF–U containing restriction enzyme site for SalI in front of nucleotides 1 bp to 20 bp upstream of the start codon of BAT2, and BAT2 ORF–D containing restriction enzyme site for PstI in front of nucleotides 1350 bp to 1331 bp of the complementary strand upstream of the start codon of BAT2, were used to clone parts of the promoter gene of BAT2 by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with SalI and PstI, and ligated into the SalI and PstI digestion sites of the plasmid YIplac211–BAT2p–PGK1p, resulting in the plasmid YIplac211–BAT2p–PGK1p–BAT2. In this study, plasmid YIplac211–BAT2p–PGK1p is used to overexpress BAT2 gene.
The plasmid was linearized by digestion with SpeI before transformation of yeast to SC minus uracil medium plates using the lithium acetate method. Correct insertion of the plasmid into the BAT2 locus on chromosome was verified by PCR. For these purpose primers BAT2 promoter–U and BAT2 ORF-D were used. Loop-out of the URA3 marker gene by homologous recombination of the two direct BAT2 promoter sequences was obtained by cultivating the correct transformants on 5–FOA plates. Correct loop-out of the URA3 gene was verified by PCR, and the primers BAT2 promoter-U and BAT2 ORF–D were used. Correct engineered strains containing PGK1p-BAT2 in place of endogenous BAT2.
Plasmid YEplac181–PGK1p–ILV2 construction
PGK1p was amplified by PCR using primers PGK1 promoter–U (containing BamHI enzyme site) and PGK1 promoter–D (containing SalI enzyme site) and inserted into SalI and BamHI sites of shuttle plasmid YEplac181, resulting in plasmid YEplac181–PGK1p.
Primer Ilv21 to 20–U containing restriction enzyme site for SalI in front of nucleotides 1 bp to 20 bp upstream of the ATG start codon of ILV2, and primer ILV2ORF–D containing the restriction enzyme site for PstI in front of nucleotides of 2559 bp to 2540 bp downstream of the ATG start codon of ILV2, were used to clone a 2559 bp fragment of ILV2 by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with SalI and PstI, and ligated into the SalI and PstI digestion sites of the plasmid YEplac181–PGK1p, resulting in plasmid YEplac181–PGK1p–ILV2. In this study, plasmid YEplac181– PGK1p–ILV2 is used to overexpress ILV2 gene. Plasmid YEplac181– PGK1p–ILV2 was also introduced into mutants using the lithium acetate method and the transformants were selected on SC minus leucine medium. By overexpressing of BAT2 and ILV2 we obtained engineered strain W303–1A YEplac181.
pUC18–GPD2P–RYUR–GPD2T plasmid construction
GPD2P–U containing restriction enzyme site for HindIII in front of nucleotides 954 bp to 935 bp upstream of the start codon of GPD2, and GPD2P–D containing restriction enzyme site for PstI in front of nucleotides 243 bp to 262 bp of the complementary strand upstream of the start codon of GPD2, were used to clone parts of the promoter gene of GPD2 promoter by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with HindIII and PstI, and ligated into the HindIII and PstI digestion sites of the shuttle plasmid pUC18–RYUR, resulting in the plasmid pUC18–GPD2–RYUR.
GPD2T–U containing restriction enzyme site for BamHI in front of nucleotides 1238 bp to 1258 bp upstream of the start codon of GPD2, and GPD2T–D containing restriction enzyme site for EcoRI in front of nucleotides 1723 bp to 1704 bp of the complementary strand upstream of the start codon of GPD2, were used to clone parts of the promoter gene of GPD2 promoter by PCR with the Pyrobest DNA polymerase (TAKARA). The fragment was digested with BamHI and EcoRI, and ligated into the BamHI and EcoRI digestion sites of the plasmid pUC18–GPD2P–RYUR, resulting in the plasmid pUC18– GPD2P– RYUR–GPD2T.
Plasmid pUC18–GPD2P–RYUR–GPD2T was digested with EcoRI and HindII I and transformed into W303–1A YEplac181 using the lithium acetate method. Correct insertion of the plasmid into the GPD2 locus on chromosome was verified by PCR. For these purpose primers GPD2P–U and GPD2T–D were used. Finally, RYUR fragment including URA3 marker was pop out by homologous recombination of the two direct Repeat sequences was obtained by cultivating the correct transformations on 5-FOA plates. Correct loop-out of the URA3 gene was verified by PCR, and the primers GPD2P–U and GPD2T–D were used. Correct engineered strain was denoted as HZAL–13 pILV2.
Plasmid pUC18–PDC6T construction
CPDC6P-U containing restriction enzyme site for EcoRI in front of nucleotides 530 bp to 511 bp upstream of the start codon of PDC6, and CPDC6P-D containing restriction enzyme site for BamHI in front of nucleotides 31 bp to 50 bp of the complementary strand upstream of the start codon of PDC6, were used to clone parts of the promoter gene of PDC6 promoter by PCR with the Pyrobest DNA polymerase (TARARA).The fragment was digested with EcoRI and BamHI , and ligated into the EcoRI and BamHI digestion sites of the shuttle plasmid pUC18-RYUR, resulting in the plasmid pUC18-PDC6P-RYUR.
CPDC6T-U containing restriction enzyme site for PstI in front of nucleotides 150 bp to 132 bp upstream of the start codon of PDC6, and CPDC6T-D containing restriction enzyme site for HindIII in front of nucleotides 2146 bp to 2127 bp of the complementary strand upstream of the start codon of PDC6, were used to clone parts of the promoter gene of PDC6 promoter by PCR with the Pyrobest DNA polymerase (TARARA). The fragment was digested with PstI and HindIII, and ligated into the PstI and HindIII digestion sites of the plasmid pUC18- PDC6P-RYUR, resulting in the plasmid pUC18-PDC6P-RYURPDC6T.
Plasmid pUC18-PDC6P-RYUR-PDC6T was linearized by digestion with EcoRI and HindIII before transformation of HZAL–13 pILV2 to SC minus uracil medium plates using the lithium acetate method. Correct insertion of the plasmid into the PDC6 locus on chromosome was verified by PCR. For these purpose primers CPDC6P-U and CPDC6T-D were used. Finally, RYUR fragment including URA3 marker was pop out by homologous recombination of the two direct Repeat sequences was obtained by cultivating the correct transformations on 5-FOA plates. Correct loop-out of the URA3 gene was verified by PCR, and the primers CPDC6P-U and CPDC6T-D were used. Correct engineered strain was denoted as HZAL–14 pILV2.
Analyses
Growth determination
Growth was followed by measuring the absorbance of the cultures at 600 nm in an INESA 721G spectrophotometer (INESA ANALYTICAL INSTRUMENT CO, LTD, China).
Measurement of glucose, isobutanol, ethanol, glycerol and acetic acid
The samples (0.002 l each) were centrifuged for 10 min at 15871 g and the resulting supernatants were frozen –20°C until analysis. Growth curves were measured using a UV–721G spectrophotometer. Concentration of glucose, ethanol, glycerol, and acetic acid concentrations were determined by a high–performance liquid chromatography (HPLC) system (Agilent Technologies 1260 Series) equipped with a Carbomix H–NP column and a refractive index (RI) detector. The column was eluted with 2.5 mM H2SO4 at a flow rate of 0.6 ml/min at 55°C, and RI detector was kept at 35°C. Isobutanol concentration was quantified by a gas chromatography (GC) system (Bruker 456-GC) equipped with a HP-INNOWAX column (length of 60 m, 0.32 mm of an inner diameter). The column temperature was controlled by 80°C for 10 min. The injector and detector temperatures were maintained at 200°C and 300°C.
Determination of dry weight
Samples (50 ml) were centrifuged at 15871 g for 10 min and washed twice with water, and subsequently the pellets were kept at 100°C for 24 h before temperature equilibration and weighing.
Results and Discussion
Growth characteristics of engineered strains
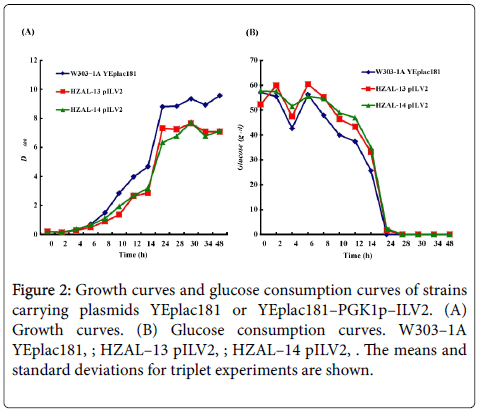
The fermentation properties of control strain (W303–1A YEplac181), HZAL–13 pILV2 and HZAL–14 pILV2 were studied under microaerobic condition. The growth rate of HZAL–13 pILV2 was similar to that of HZAl–14 pILV2 (Figure 2a). However, the growth rate of the two-engineered strains HZAL–13 pILV2 and HZAL–14 pILV2 were both slower than that of control strain. These results were further more supported by the fact that the consumption rates of glucose in HZAL–13 pILV2 and HZAL–14 pILV2 lagged a little compared to control strain (Figure 2b). Finally, the biomass concentrations in HZAL–13 pILV2 and HZAL–14 pILV2 at the end of the growth period were similar but both less than the control strain (Table 4).
| Strains | Biomass concentration (g dl-1) |
|---|---|
| W303–1A YEplac181 | 0.121 ± 0.0012 |
| HZAL–13 pILV2 | 0.1022 ± 0.0010 |
| HZAL–14 pILV2 | 0.0924 ±0.0009 |
Table 4: Biomass concentration of strains.
Isobutanol titer
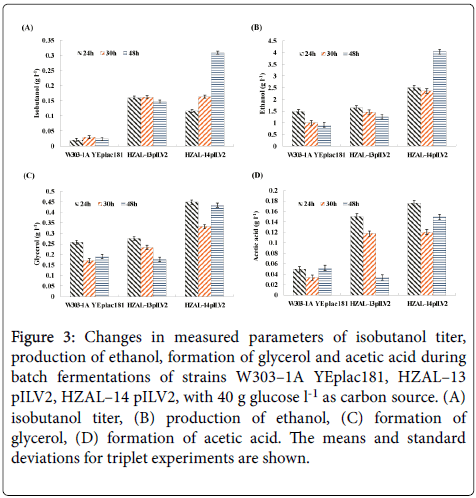
Our results showed that isobutanol titers of HZAL–13 pILV2 and HZAL–14 pILV2 were both increased during microaerobic fermentations compared with control strain (Figure 3a). The maximum isobutanol titer of control strain was 29.8 mg/l. While HZAL–13 pILV2 exhibited a 445 % increase in isobutanol titer (162.3 mg/l) compared with control strain. Meanwhile the maximum isobutanol yield of HZAL–14 pILV2 was increased 938 % (309.3 mg/l) than that of control strain. One explanation is that GPD2 is the key gene to synthesize glycerol [20,21], deletion of GPD2 could decrease glycerol formation and help to further enhance the metabolic fluxes leading to isobutanol production. It was reported that overexpression of BAT2 would increase more carbon source flow to isobutanol biosynthesis direction in S. cerevisiae [22-24]. It was also reported that deletion of PDC1 would increase isobutanol production in S. cerevisiae [25]. By deleting PDC6 and GPD2, parts of carbon source flux that would be used to form ethanol and glycerol has been used to synthesize isobutanol during microaerobic fermentations in HZAL–14 pILV2. Therefore, the integrative effect in isobutanol production was obtained with BAT2 and ILV2 overexpression and GPD2 and PDC6 deletion.
Production of ethanol
The maximum ethanol production of control strain (W303–1A YEplac181), HZAL–13 pILV2 and HZAL–14 pILV2 were 1481.4 mg/l, 1641.7 mg/l and 4040.9 mg/l, respectively (Figure 3b). Ethanol is the main product in S. cerevisiae [26]. This is the reason why ethanol production level was slightly increased after deleting GPD2 in HZAL– 13 pILV2 than control strain. However, HZAL–14 pILV2 obtained a higher ethanol production compared with control strain. In addition to PDC6, PDC5 and PDC1 coding of pyruvate decarboxylase can also be used for catalyzing pyruvate into acetaldehyde. And Pdc5 and Pdc1 play major roles during ethanol fermentation [27,28]. The ethanol titer of HZAL–14 pILV2 was increased may probably due to enhancement of the functions of Pdc5 and Pdc1 after deleting GPD2 and PDC6 or any other reason. The precise molecular mechanisms need to be further investigated.
Formation of glycerol and acetic acid
We also investigated the effect of GPD2 and PDC6 deletion on the formation of other secondary metabolic products such as glycerol and acetic acid (Figures 3c-3d). The maximum glycerol formation of control strain, HZAL–13 pILV2 and HZAL–14 pILV2 were 257.7 mg/l, 275.5 mg/l and 448.8 mg/l, respectively. And the maximum acetic acid formation of control strain, HZAL–13 pILV2 and HZAL–14 pILV2 were 51.8 mg/l, 150.3 mg/l and 175.8 mg/l, respectively. The glycerol formation of HZAL–13 pILV2 was similar to that of control strain. However, the glycerol formation of HZAL–14 pILV2 increased 74% than that of control strain. On the other hand, acetic acid formation in HZAL–13 pILV2 and HZAL–14 pILV2 showed 190% increase and 239% increase, respectively, compared with the control strain.
Isobutanol exhibits superior physicochemical properties as an alternative biofuel and can be transported using current petroleum pipelines [3]. However, ethanol and glycerol are major unwanted byproducts in isobutanol fermentation in S. cerevisiae [26]. To increase isobutanol titers in S. cerevisiae, two strains were obtained by deleting GPD2 and PDC6, and simultaneous overexpressing of BAT2 in S. cerevisiae. Microaerobic fermentations showed that the maximum isobutanol titers of HZAL-13 pILV2 and HZAL-14 pILV2 were 162.3 mg/l and 309.3 mg/l, respectively. Compared with control strain, isobutanol titers of HZAL-13 pILV2 and HZAL-14 pILV2 were increased 445 % and 938 %, respectively. In this study, only ILV2 (encoding acetolactate synthase which catalyze the first step in valine synthesis) was overexpressed. Isobutanol might further improved by simultaneously overexpressing ILV3 and IVL5.
Biomasses of HZAL–13 pILV2 and HZAL–14 pILV2 decreased markedly than that of control strain. Isobutanol titer of HZAL–14 pILV2 increased dramatically than that of control strain. While ethanol titer of HZAL–14 pILV2 increased markedly than that of control strain. These results are unexpected. One explanation perhaps is that GPD1, PDC5 and PDC1 still play important roles in HZAL–14 pILV2. Higher acetic acid titers in HZAL–13 pILV2 and HZAL–14 pILV2 perhaps to solve cofactors imbalance.
So far, there are many studies on improving isobutanol titers in S. cerevisiae. The accumulation of alcohols including isobutanol and ethanol is toxic to S.cerevisiae, which has limited the production capacity of many yeast strains in fermentation [26,29]. For this problem, investigation on improving isobutanol titer by adding potassium and hydroxide ions to the medium in which yeast grow has been reported recently [30]. There are also many other strategies. One strategy has been used for construction of higher isobutanolproducing titer strains of S. cerevisiae by re-locating the valine biosynthesis enzymes ILVs from the mitochondrial matrix into the cytosol [12]. Other strategies are overexpression or deletion of genes in isobutanol biosynthesis in S. cerevisiae. For example, overexpression of ILVs and BAT2 to increase flux in the isobutanol biosynthetic pathway [1]; deletion of BAT1 [10] or PDC1 [25] to eliminate competing pathways. But there have been no strategies about improving isobutanol titer by deleting GPD2 gene in S. cerevisiae. This is the first report about improving isobutanol titer by deletion of GPD2 and simultaneous deletion of PDC6 in S. cerevisiae. And this will provide guidance for future studies about improving isobutanol production.
In summary, the present study has demonstrated the proposed concept to increase the isobutanol titer by decreasing glycerol formation and ethanol biosynthesis in combination through deletion of PDC6 and GPD2 is feasible in S. cerevisiae.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21206028), co-financed by Doctoral Fund of Ministry of Education (No. 20121317120014), co-financed by the Hebei Province Natural Science Fund (No. B2013202288), co-financed by the Hebei Provincial Office of Education Science and Technology Research Projects (N0. q2012024), co-financed by the Hebei University of Technology Outstanding Youth Science and Technology Innovation Fund (No. 2012009), co-financed by the Open Fund of Key Laboratory of System Bioengineering of Ministry of Education (Tianjin University) (No. 20130315).
Conflict of Interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
References
- Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K (2011) Increased isobutanol production in Saccharomyces cerevisiaeby overexpression of genes in valine metabolism. Biotechnol Biofuels 4: 2089-2090.
- Shi A, Zhu X, Lu J, Zhang X, Ma Y (2013) Activating transhydrogenase and NAD kinase in combination for improving isobutanol production.MetabEng 16: 1-10.
- Su H, Zhao Y, Zhao H, Wang M, Li Q, et al. (2014) Identification and assessment of the effects of yeast decarboxylases expressed in Escherichia coli for producing higher alcohols.J ApplMicrobiol 117: 126-138.
- Matsuda F, Ishii J, Kondo T, Ida K, Tezuka H, et al. (2013) Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance. Microb Cell Fact 12: 119.
- Qi H, Li S, Zhao S, Huang D, Xia M, et al. (2014) Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis. PloS One 9: e93815.
- Connor MR, Liao JC (2009) Microbial production of advanced transportation fuels in non-natural hosts.CurrOpinBiotechnol 20: 307-315.
- Weber C, Farwick A, Benisch F, Brat D, Dietz H, et al. (2010) Trends and challenges in the microbial production of lignocellulosicbioalcohol fuels.ApplMicrobiolBiotechnol 87: 1303-1315.
- Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae.MicrobiolMolBiol Rev 72: 379-412.
- Porro D, Gasser B, Fossati T, Maurer M, Branduardi P, et al. (2011) Production of recombinant proteins and metabolites in yeasts: when are these systems better than bacterial production systems?ApplMicrobiolBiotechnol 89: 939-948.
- Park SH, Kim S, Hahn JS (2014) Metabolic engineering of Saccharomyces cerevisiae for the production of isobutanol and 3-methyl-1-butanol.ApplMicrobiolBiotechnol 98: 9139-9147.
- Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74: 2259-2266.
- Brat D, Weber C, Lorenzen W, Bode HB, Boles (2012) Cytosolic re-localization and optimization of valine synthesis and catabolism enables inseased isobutanol production with the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 5: 65.
- Velasco JA, Cansado J, Peña MC, Kawakami T, Laborda J, et al. (1993) Cloning of the dihydroxyaciddehydratase-encoding gene (ILV3) from Saccharomyces cerevisiae.Gene 137: 179-185.
- Yu KO, Kim SW, Han SO (2010) Reduction of glycerol production to improve ethanol titer in an engineered Saccharomyces cerevisiae using glycerol as a substrate. J Biotechnol 150: 209-214.
- Thomas BJ, Rothstein R (1989) The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene.Genetics 123: 725-738.
- Burke D, Dawson D, Stearns T (2000) Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. CSHL Press 174.
- Sambrook J, Fritsch EF, Maniatis T (1989). Molecular cloning: a laboratory manual. New York: Cold spring harbor laboratory press.
- Gietz RD, Sugino A (1988) New yeast Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 525-534
- Zhang AL (2005) Optimization of ethanol production in Saccharomyces cerevisiae by genetic engineering of minimizing glycerol biosynthesis. Tianjin University.
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts.MicrobiolMolBiol Rev 66: 300-372.
- Kong QX, Zhang AL, Cao LM, Chen X (2007) Over-expressing GLT1 in a gpd2Delta mutant of Saccharomyces cerevisiae to improve ethanol production.ApplMicrobiolBiotechnol 75: 1361-1366.
- Eden A, Van Nedervelde L, Drukker M, Benvenisty N, Debourg A (2001) Involvement of branched-chain amino acid aminotransferases in the production of fuel alcohols during fermentation in yeast.ApplMicrobiolBiotechnol 55: 296-300.
- Yoshimoto H, Fukushige T, Yonezawa T, Sone, H (2002) Genetic and physiological analysis of branched-chain alcohols and isoamyl acetate production in Saccharomyces cerevisiae. ApplMicrobiolBiotechnol 59: 501-508.
- Zhang CY, Qi YN, Ma HX, Li W, Dai LH, et al. (2015) Decreased production of higher alcohols by Saccharomyces cerevisiaefor Chinese rice wine fermentation by deletion of Bat aminotransferases.J IndMicrobiolBiotechnol 42: 617-625.
- Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, et al. (2012) Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol 159: 32-37.
- Generoso WC1, Schadeweg V, Oreb M, Boles E (2015) Metabolic engineering of Saccharomycescerevisiae for production of butanol isomers.CurrOpinBiotechnol 33: 1-7.
- Hohmann S (1991) PDC6, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC1 promoter.Curr Genet 20: 373-378.
- Agarwal PK, Uppada V, Noronha SB (2013) Comparison of pyruvate decarboxylases from Saccharomyces cerevisiae and Komagataellapastoris(Pichiapastoris).ApplMicrobiolBiotechnol 97: 9439-9449.
- Lam FH, Ghaderi A, Fink GR, Stephanopoulos G (2014) Biofuels. Engineering alcohol tolerance in yeast.Science 346: 71-75.
- Trafton A (2014) New approach to boosting biofuel production. MIT News Office.
Copyright: © 2016 Zhang A et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.