Research Article - (2018) Volume 4, Issue 3
Efficacy of Facultative Oligotrophic Bacterial Strains as Plant Growth-Promoting Rhizobacteria (PGPR) and their Potency Against Two Pathogenic Fungi Causing Damping-off Diseases
*Corresponding Author: Sayed Abdelaziz, Department of Agricultural Microbiology, Faculty of Agriculture, Fayoum University, Egypt Email:
Abstract
Plant growth promoting is a multitask phenomenon. The bacteria achieve this by suppressing plant pathogens, production of plant-growth regulators, fixing atmospheric nitrogen, and solubilizing phosphate and micronutrients. Therefore, the present study aimed to examine the facultative oligotrophic bacteria isolates as biocontrol and plant growth promoting agents. The test isolates (FNS1 and FNS2) possessed Plant Growth Promoting Rhizobacteria (PGPR) (abilities including Indole Acetic Acid (IAA) production, Salicylic acid (SA) production, zinc and phosphate solubilization, N2-fixation, cellulase and chitinase production) and potency against pathogenic fungi causing damping-off disease (Pythium ultimum and Rhizoctonia solani). Isolates were tested against high temperature, pH and salt tolerant characters which prevail in this region. Molecular identification by 16s rDNA and nucleotide sequencing was made for the test isolates. Snap bean (Phasolius vulgaris L. cv, Xera) plants were inoculated with bacterial isolates (FNS1 and FNS2) in presence of 1/3 Nitrogen (N)-recommended dose. Inoculated plants increased significantly in N-content and plants dry weight. The antagonizer strains together enhanced the level of defense principles like Peroxidase (PO), Polyphenol Oxidase (PPO) and Total phenol content of plants. Damping-off disease suppression was expressed as there was increase in the percentage of survived plants which were inoculated when compared with uninoculated plants. These results indicate that Bacillus thioparans and Stenotrophomonas maltophilia have growth promoting potential and can be tested at field level for investment as bioinoculant.
Keywords: PGPR; Oligotrophic bacteria; Putative N2-fixers; Biocontrol agents; 16s rDNA; Phylogenetic analysis
Introduction
The rhizosphere is defined as the soil around and in contact with plant root surface, which is rich in nutrient and microorganism activity, characterized by complex and dynamic physical, chemical and biological interactions. In the rhizosphere, microorganisms have a great role in the organic matter transformations and biogeochemical cycles of plant nutrients. An essential number of bacterial species interact with their host plants and may extend beneficial impacts on plant growth, plant nutrition and disease suppression [1,2].
Plant growth promoting is a multitask phenomenon. The bacteria achieve this by suppressing plant pathogens, production of plant-growth regulators, fixing atmospheric nitrogen, and solubilize phosphate and micronutrients, production of phytohormones and enzymes, etc. Nowadays, PGPR represent an attractive alternative for the overuse of harmful agrochemicals [3]. Beside, PGPR are able to ameliorate abiotic and/or biotic stressors, which can be exploited to promote plant growth and productivity of the plants under stress conditions [4].
Specific studies showed the role of PGPR strains based on disease suppression, PGPR have been identified as a biological control alternative to pesticide use by achieving disease suppression without negative effects on user, consumer or the environment. They are capable of suppressing microorganisms or nematodes that cause root damage; also they have a role of defense enzymes like Phenylalanine Ammonia Lyase (PAL), PO and PPO. The earlier studies evinced that PGPR can be used as an effective bio inoculants for plant growth promotion and controlling the wide range of plant pathogenic fungi, therefore have great potential for biotechnology applications [5-7].
This study was an attempt to use selective strains of facultative oligotrophic bacteria (FNS1 and FNS2) that have broad spectrum of plant growth-promoting abilities and antagonistic potential against phytopathogenic fungi which could be used as safe alternative for the overuse of harmful agrochemicals. For this we chose the most effective facultative oligotrophic bacterial strains obtained from our previous study which examined abilities like IAA production, SA production, zinc and phosphate solubilization, N2-fixation, cellulase and chitinase production [8]. It is worth to mention that two strains used in this study as PGPR were isolated from the same soil at Fayoum Governorate and were identified as Bacillus thioparans and Sterotrophomons maltophilia with accession number MH000622 and MH000623 respectively. The effect of these two strains on growth of snap bean crop plant (Phasolius vilgares) can be well observed, when added separately or together as PGPR or when added to control damping-off disease caused by Pythium ultimum and Rhizoctonia solani.
Materials and Methods
Isolation, morphological, physiological and biochemical characters of oligotrophic isolates
The bacterial isolates (FNS1 and FNS2) were isolated from the deficient soil in Fayoum governorate, using nutrient and diluted nutrient broth medium (NB and DNB media). Different morphological characters of obtained isolates were observed on NB agar media incubated for two weeks at 30°C. Cell shape was examined microscopically using gram stain in addition to sporulation and motility. The production of acids from selected sugars, solubilization of phosphate [9], Zinc [10] was carried out. The capability to produce Plant Growth Promoting substances (PGPs) such as IAA [11], SA [12], Siderophores production [13], cellulase production [14] chitinase production [15] and N2-fixation [16] was detected. Isolates were screened in vitro for antagonism towards soil borne pathogenic fungi: Pythium ultimum and Rhizoctonia solani, in dual culture test [17]. Antagonistic activity was assessed by relating mycelial diameter on plates inoculated with bacteria to mycelial diameter on control plates (plates inoculated with target fungi alone) and computing percentage inhibition. The isolates were examined against some adverse environmental conditions to determine their capability to live, proliferate sustain life and perform their activities. They were tested against temperature, increasing pH and salt tolerant characters which prevail in this region.
Extraction of DNA, PCR amplification and sequence analysis of 16S rDNA gene
Genomic DNA extraction from bacterial isolates (FNS1 and FNS2) was done by using DNA extraction protocol according to Moore et al. [18]. 16S rDNA sequence was amplified from genomic DNA obtained by PCR with the U1492R primer: 5′-GGT TAC CTT GTT ACG ACT T-3′ and the 8F primer: 5′-AGA GTT TGA TCC TGG CTC AG-3′ [19]. PCR reactions were performed in a total volume of 50 μl, containing 100 ng of the template DNA, 0.2 µM concentration of each primer, 200 µM concentration of each dNTPs and 2.5 U of Taq polymerse enzyme, 10x PCR buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl) and water nuclease-free. PCR amplification was performed in a thermal cycler 2720 (Applied Biosystems, USA). The amplified PCR product was purified using Wizard RSV gel and PCR clean-up system (Promega catalog number A928) (50 reps), following manufacture instructions to remove unincorporated PCR primers. The purified PCR products were subjected to sequencing through Solgent Sequencing Service located in Korea. The sequences of ITS region of the two bacteria isolates were submitted to GenBank under the following accession numbers (MH000622 and MH000623) for Bacillus thioparans and Stenotrophomonas maltophilia (FNS1 and FNS2) respectively.
Computational analysis (blast) and construction of phylogenetic tree
The data of the nucleotide sequence of the ITS regions of rDNA obtained from the two isolates were compared with ITS sequences collected from Bacillus thioparans and Stenotrophomonas maltophilia sequences available in GenBank database (http://www.ncbi.nlm.nih.gov/Blast). The nucleotide sequences were aligned using Clustal W 2.1 multiple sequence alignment [20]. Phylogenetic dendrogram was constructed by the Clustal W 2.1 multiple sequence alignment programs. Unweighted Pair Group Method with Arithmetic Mean (UPGMA) was used for creation of rooted phylogenetic tree.
Preparation of facultative oligotrophic bacterial inoculants
Inoculants for treatment of seeds were prepared by streaking isolates (FNS1 and FNS2) individually onto NB agar plates. After incubating plates at 28ºC for 48 h, bacterial cells were harvested off plates by drenching 10 ml sterile 0.1 M MgSO4 solution (pH 7.0) and scrapping with a sterile glass spreader. The resulted cell suspension was collected in sterile flask, diluted with sterile 0.1 M MgSO4 solution to give cell concentration of ca. 108 ml-1 then supplemented with Carboxymethylcellulose (CMC) to a 1% concentration. The mixture inoculant was prepared by mixing equal volumes of cell suspensions of individual strain.
Fungal inoculum (pathogenic agents)
For preparation of fungal inoculums (used as pathogenic agents), the two selected strains Py. ultimum and R. solani were grown in sterile sand (50 g), maize meal (1.5 g), water (10 ml) medium in 300 ml Erlenmeyer flask for 3 weeks at 28°C in the dark .The fungal inoculums was incorporated into the potting mix prior to planting seeds or seedlings at the rate of 0.5% (wt/vol). None infested control pots were treated with fungal inoculums [21].
Seeds preparing
Snap bean seeds were surface disinfected by dipping them is 2% sodium hypochlorite for 30 s followed by washing in Sterile Distilled Water (SDW), dipping them again in 70% ethanol for 30 s and rinsing in five changes of SDW [22].
The surface-disinfected seeds were prepared by inoculation with antagonizers suspensions by repeated emersion in stock culture of antagonizer organisms.
Root colonization assay and antibiosis between facultative oligotrophic bacterial strains selected for bioassay experiments
Sterilized glass test tubes were filled with NB agar medium to a depth of 6 cm. One bacterial treated seed or control seed was placed on the surface of the medium. Tubes were sealed with parafilm; the lower part was wrapped with aluminum foil, and incubated at room temperature (25 ± 3°C). After seedling roots reach the end of the agar, seedlings were aseptically removed from the tubes; root were sliced into segments and placed onto NB agar medium plates, three plates were used for each isolate. The plates were incubated at 28°C for 3 days and examined for signs of bacterial growth around the root segments indicating root colonization with tested bacteria.
To ensure from the compatibility of the isolates (FNS1 and FNS2) when applied in mixture, the isolates were individually tested for in vitro antibiosis against all other strains. The test was performed using a cross-streak method in which NB agar medium plates were inoculated with a single isolate as streak across the surface of plate and incubated at 28ºC for 48 h [23]. The other isolate was cross-streaked perpendicular to the growth streak of the first isolate and incubated again at 28ºC. After 48 h, plates were observed for inhibition of growth at each side of the first isolate.
Seed germination assay
Theisolates (FNS1 and FNS2) were tested individually and in mixtures for their effects on seed germination and seedling vigor of snap bean plants. The surface-disinfected seeds were placed on surface of sterile water agar (0.75%) plates and treated with bacterial suspension by dropping 30 µl seed-1. Three replication plates with 25 seeds per plate were used for each strain. Seeds in control plates were treated with 30 µl seed-1 sterile MgSO4 solution and all plates incubated at room temperature. After 10 days, number of germinated seeds and seedling length (shoot+root) were recorded from which the germination percentage and vigor index were calculated. The following formula of Abdul Baki and Anderson was used to calculate the Vigor index:Vigor index=percent germination × seedling length [24].
Layout of the greenhouse experiment
The layout of experiment showed in Table 1. The air dried soil used in this experiment was crushed and sieved through 2 ml sieve, the soil characterizations was studied as shown in Table 2. Soil was sterilized by autoclaving at 121°C for 1 h. Pots with 14 cm width was used for planting and was prepared by washing with water repeatedly then surface sterilizing with 70% ethyl alcohol. Sterilized soil was distributed and packed in plastic pots at 2.5 kg/pot. As to the potassium and phosphorus fertilizers supplementation for snap bean (Phasolius vulgaris L.) cv. Xera plant they were added at the recommended doses, 500 mgsuperphosphate (15% P2O5) and 125 mg potassium sulphate (48% K2O)/pot respectively.
| Complete dose of N, P and K | Control |
|---|---|
| 1/3dose of N, +P and K | +FNS1 |
| 1/3dose of N, +P and K | +FNS2 |
| 1/3dose of N, +P and K | +mixture of FNS1 and FNS2 |
| Complete dose of N, P and K | +Py. altimum |
| 1/3dose of N, +P and K | +Py. Altimum+FNS1 |
| 1/3dose of N, +P and K | +Py. Altimum+FNS2 |
| 1/3dose of N, +P and K | +Py. Altimum+mixture of FNS1and FNS2 |
| Complete dose of N, P and K | +R. solani |
| 1/3dose of N, +P and K | +R. solani+FNS1 |
| 1/3dose of N, +P and K | + R. solani+FNS2 |
| 1/3dose of N, +P and K | + R. solani+mixture of FNS1and FNS2 |
FNS2 Stenotrophomonas maltophilia
Mix Mixture of FNS1 and FNS2
Table 1: The layout of greenhouse experiment.
| Soil Properties | Value |
|---|---|
| Texture class | Sandy Loam |
| Calcium carbonates% | 8.9 |
| pH (in soil paste) | 7.66 |
| ECe (dS/m) | 2.89 |
| Organice matter Contents% | 0.90 |
| Total Nitrogen (mg/kg soil) | 12.8 |
| Available Phosphorus (mg/kg soil) | 4.25 |
| Available Potassium (mg/kg soil) | 22.57 |
| DTPA extractable-Fe (mg/kg soil) | 2.45 |
| DTPA extractable-Mn (mg/kg soil) | 2.28 |
| DTPA extractable-Zn (mg/kg soil) | 0.76 |
| DTPA extractable-Cu (mg/kg soil) | 0.14 |
Table 2: Chemical and physical properties of the soil.
Notice: The recommended doses of potassium and phosphorus fertilizers for snap bean (Phasolius vulgaris L.) cv. Xera, were 200 kg superphosphate (15% P2O5) and 50 kg potassium sulphate (48% K2O)/feddan respectively. N-recommended dose (80 unit N2/feddan equal 240 kg ammonium sulphate, 33%N2/feddan) added at four times.
The greenhouse experiment
Ten surface-disinfected seeds inoculated with isolates (FNS1 and FNS2) were planted in 14 cm diameter pots filled with sterilized soil infested or not infested with one of the two fungi. The fungal inocula were incorporated into pots at the rate of 0.5 (w/v) and saturated with water. After planting, the surface of each pot was covered with a 2 cm layer of sterilized sand. Before the sand cover, each hill was supplemented with additional 1 ml the appropriate antagonizer inoculum. Soil moisture was raised to 70% of its water holding capacity at the beginning of the experiment. After 8 days of planting, pre damping-off disease incidence was recorded which was indicated by complete absence of germination of the plant from soil surface. While the post damping-off was recorded after 15 days of planting and the number of dead plants, after germination, was calculated. After fifteen days of planting, plants were thinned to five only (snap bean) per pot. Then the first dose of N fertilizer was added (20 kg ammonium sulphate, 33% N/feddan equal 50 mg/pot), the second dose of N fertilizer (20 kg ammonium sulphate, 33% N/feddan equal 50 mg/pot) was added after two weeks of planting. All plants were harvested at 30 days age. After 30 days of planting, plants were carefully removed from pots, washed with distilled water to get rid of soil particles, and then dried with paper towel to eliminate any excess or dust. Morphological estimates were limited to the determination of whole plant dry weight and percentage of survived plants from pre and post damping-off disease. All collected plants in all treatments were kept and prepared for analysis.
Chemical, enzymatical determinations:
Total nitrogen was determined according to the well-known method described by Donald et al. [25]. Estimation of total phenols, PO and PPO were carried out by the method described by Ramamoorthy et al. [26].
Statistical analysis
Data were statistically analyzed by Analysis of Variance (ANOVA), and mean separated by Duncan’s Multiple Range test at a significance level of P ≤ 0.05.
Results
Morphological, physiological and biochemical characters of oligotrophic isolates
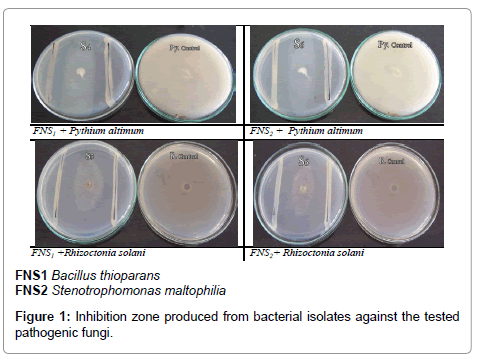
The possible morphological and physiological characters of the isolates were summarized in Table 3; the capability of isolates to produce some plant growth promoters was also determined. The ability of isolates to grow on N-free media (Putative N2 fixers) was also examined in suitable liquid culture media. The isolates capable of growing in medium free of any N-source for five successive subculture was considered as putative N2 fixer whether the fixation was via Nirogenase or by scavenging nitrogen from the surrounding. Both isolates (FNS1 and FNS2) were positive for indole, SA production, phosphate and zinc solubilization, nitrogen fixation, cellulose and chitin hydrolysis and were tolerant for up to 15% NaCl, pH tolerance was found in a range of pH 6-12, also the isolates showed growth at high temperature (up to 50°C). They also exhibited antifungal activity against Pythium ultimum and Rhizoctonia solani (Figure 1). The strains had the same efficiency whether in extreme or in normal conditions.
| Characteristic/strain | B. thioparans MH000622 | S. maltophillia MH000623 |
|---|---|---|
| Gram stain | + | - |
| Motility | + | + |
| Spore forming | + | - |
| Growth at | ||
| 10,20,30,40 and 50oC | + | + |
| Indole production | + | - |
| Citrate utilization | + | - |
| Fermentation of | ||
| D-glucose | + | + |
| Lactose | - | + |
| D-mannitol | + | - |
| D-xylose | + | + |
| Growth in NaCl | ||
| 5% | + | + |
| 10% | + | + |
| 15% | + | + |
| pH 6, 8, 10 and 12 | + | + |
| Metabolites and hydrolytic Enzymes | ||
| Oxidase | + | + |
| Catalase | + | - |
| Gelatin hydrolysis | + | + |
| Starch hydrolysis | + | - |
| Cellulose hydrolysis | + | + |
| Chitinase | + | + |
| Activities related to plant growth promotion | ||
| Siderophore production | + | + |
| Indoleacetic acid production | + | + |
| Salyselic acid production | + | + |
| Phosphate solubilization | + | + |
| Zinc solubilization | + | + |
| Putative nitrogen fixation | + | + |
| Antifungal activity (inhibition zone mm) | ||
| Py. altimum | 16.5 | 20 |
| R. solani | 15.0 | 18 |
Table 3: Morphological, physiological and biochemical characters.
Molecular identification by rDNA and nucleotide sequencing:
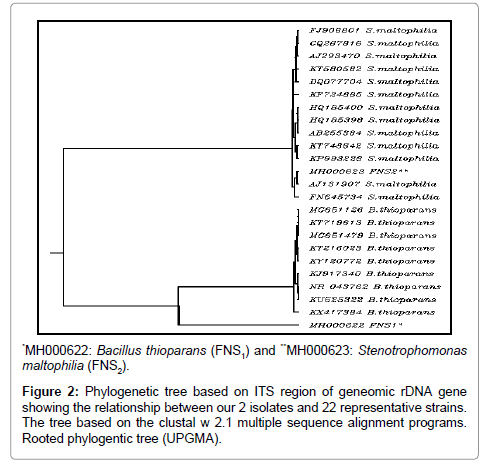
The ribosomal DNA genes (rDNA) possess characteristics that are suitable for the identification of bacteria isolates at the species level. 16S gene amplicons, encompassing hyper variable regions, used to infer taxonomic identifications based upon bioinformatics alignments against sequence databases [27-29]. The phylogenetic tree obtained by sequence analysis of ITS region of two plant growth promoting bacterial (Bacillus thioparans and Stenotrophomonas maltophilia) strains and the sequences of 22 isolates obtained from sequence databanks is represented in Figure 2. The dendrogram of bacteria isolates were shown in Figure 2. The dendrogram was divided into two clusters; the first cluster included Bacillus thioparans isolates, while the second cluster included Stenotrophomonas maltophilia isolates. The GenBank accession number for ITS sequence for our isolates is MH000622 and MH000623 for Bacillus thioparans (FNS1) and Stenotrophomonas maltophilia (FNS2) respectively.
Evaluation of facultative oligotrophic bacterial strains as PGPR inoculants
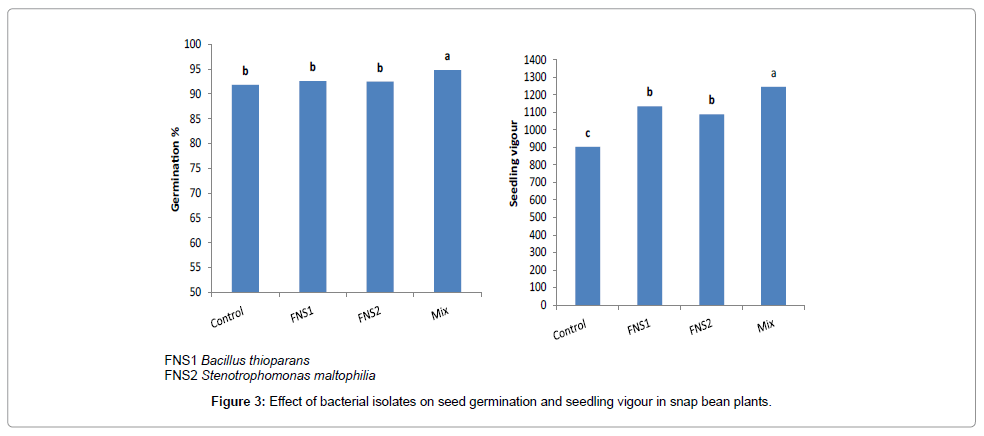
Theisolates (FNS1 and FNS2) were individually tested in vitro for antibiosis against other. However, no antibiosis was observed among strains of each mixture. Also, these strains were tested for colonization roots of the indicator plants and the test indicated that all strains were able to colonize roots. The results of seed germination experiment are presented in Figure 3. The two strains were shown to have different effects on percentage of seed germination (G%) and seedling vigor index (VI( . Inoculation with strains in mixture (Mix) only resulted in a significant increase in G% of snap bean seeds compared to control seeds. Compared to control, vigor index (VI) of snap bean seedlings was significantly increased by inoculation with (FNS1, FNS2 and Mix) respectively.
Effect of the facultative oligotrophic bacterial putative N2-fixers on growth and N-content of the experimented plant
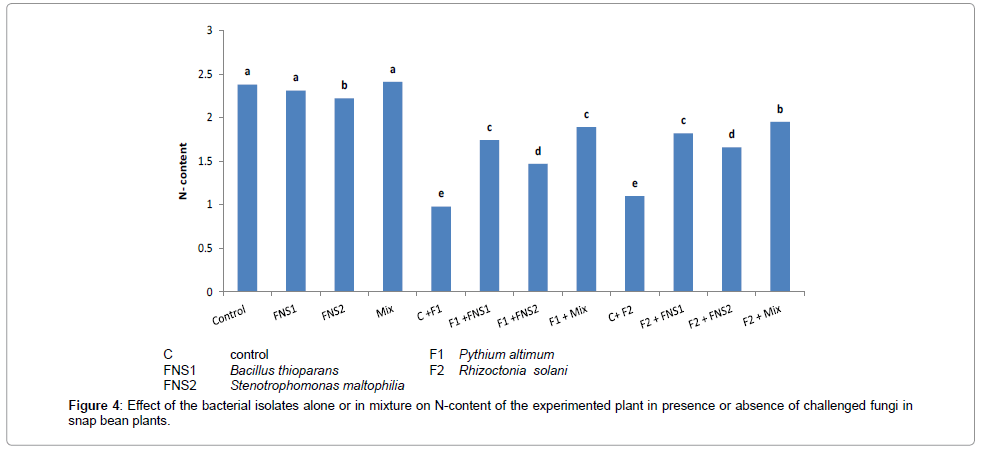
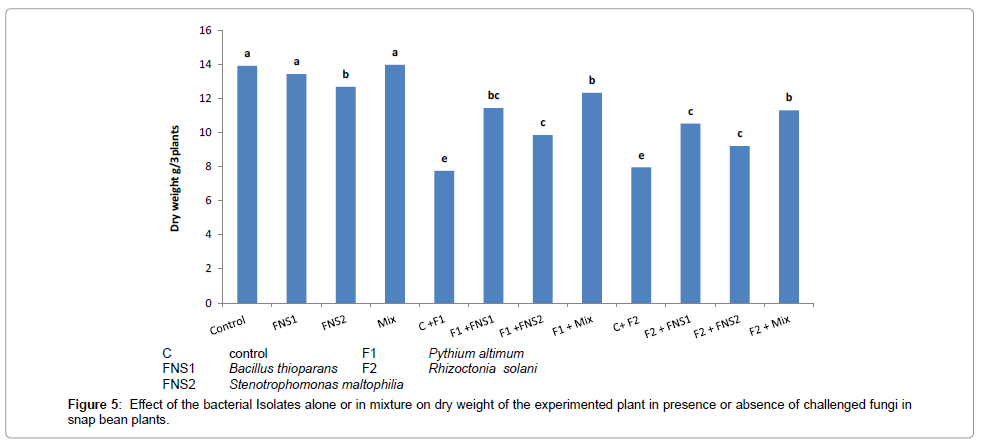
Figures 4 and 5 illustrate the effect of supplementing the plants (snap bean) with mineral nitrogen fertilizer at recommended dose (C treatment), compared with the treatments which received 1/3 recommended dose of nitrogen plus the putative nitrogen fixers strains individually or in mixed inoculants, on nitrogen content and the plant dry weight obtained at the end of the experiment (30 days). There were no significant differences between values of total nitrogen contents and total dry weight of snap bean plants. The 1/3 dose nitrogen fertilizer in presence of the mixed inoculum of the putative N2-fixers gave high significant values of nitrogen contents and dry weight of snap bean plants which were very near to that of the control (complete dose treatment C).
When the plants were challenged with the two pathogenic fungi Pythium ultimum and Rhiaocotina solani, significant decreases in nitrogen content and dry weight were observed in the control due to the weak growth of the infested plants in these treatments. When the plants were supplemented with the putative N2-fixers in presence of 1/3 nitrogen dose and then challenged with infested fungi, the adverse effect of the fungi was partially alleviated to some degrees and there were considerable increase in total nitrogen and total dry weight of the plant. The results indicated that supplementing the plants with 1/3 nitrogen fertilizer dose in presence of the putative oligotrophic N2-fixers may have alleviated the adverse effect of the pathogenic fungi on N-content and plant dry weight yield of the plant.
Facultative oligotrophic bacteria as biocontrol agents
Snap bean plants were treated with the putative N2-fixers antagonizes bacterial strains and their mixtures. Significant increases in nitrogen content and dry weight of plants was observed which may be due to the production of growth promoting substances by these strains or the increase in the defense mechanisms of the plants due to the inoculation with bacterial strains that led to increases in PO, PPO and total phenol content of plants. All bacterial strains treatments produce these compounds in significant amounts when compared with control (Table 4).
| Treatment | PO nm min/g |
PPO nm min/g |
Total phenol mg/g |
|---|---|---|---|
| Control | 0.124e | 0.142e | 0.945bc |
| FNS1 | 0.169d | 0.216d | 1.211b |
| FNS2 | 0.201d | 0.238d | 1.290b |
| Mix | 0.218d | 0.262d | 1.381b |
| Control + Py. altimum | 0.303c | 0.389c | 1.552ab |
| FNS1 + Py. altimum | 0.411a | 0.436b | 1.635ab |
| FNS2 + Py. altimum | 0.423a | 0.486a | 1.679ab |
| Mix + Py. altimum | 0.436a | 0.499a | 1.956a |
| Control + R. solani | 0.291c | 0.312c | 1.496b |
| FNS1 + R. solani | 0.394bc | 0.409b | 1.668ab |
| FNS2 + R. solani | 0.365c | 0.379b | 1.539ab |
| Mix + R. solani | 0.404b | 0.427b | 1.981a |
| LSD | 0.031 | 0.028 | 0.48 |
FNS2 Stenotrophomonas maltophilia; Mix Mixture of FNS1 and FNS2.
Table 4: Effect of bacterial isolates on, PPO, PO activities and total phenol contents in presence or absence of challenged fungi in snap bean plants.
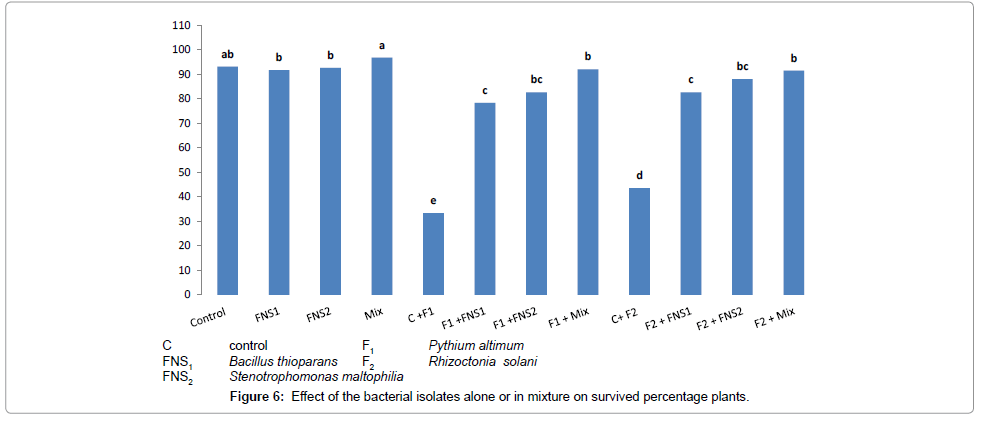
The deleterious effects due to the infestation of snap bean plants with the two pathogenic fungi resulted in little increases in values of the defense mechanism principles (PO, PPO and phenol content), and consequently gave relatively higher percentages of damping-off snap bean plants, when compared with the damping-off plants in treatments supplied with the antagonizing bacterial strains only. The plant rose its defense mechanism, when different bacterial strains were added in presence of 1/3 N-fertilize, treatment (1/3 N+mix) was highly significant when compared with the control. The PO, PPO and total phenol were increasing resistance of plants due to the presence of the mixed bacterial strains used as fungi antagonizers. Damping-off disease suppression was expressed as increase in the percentage of survival plants compared with uninoculated plants. All isolates used singly or in mixture significantly increased percentage of survival plants in presence of both fungi, Pythium ultimum and Rhizoctonia solani compared with fungi only controls as shown in Figure 6.
Discussion
The present work is an attempt to benefit from facultative oligotrophic bacterial strains isolated from poor soils prevailing in Fayoum Governorate. The isolates obtained from previous study [8] were examined and also chosen for different characters (IAA production, SA production, zinc and phosphate solubilization, siderophores production, N2-fixation, cellulase and chitinase production), also the strains were capable of growing at high temperature (50°C) and pH 12.0 producing observable growth in presence of 15% of salt. The strains had the same efficiency whether in extreme or normal conditions; they were used in this study. The strains were completely identified as Bacillus thioparans and Sterophomons maltophilia with accession number MH000622 and MH000623 respectively. The effect of these strains on growth of snap bean (Phasolius vulgaris L.) cv. Xera were studied when added separately or together as PGPR or when added to control damping-off diseases of the previously mentioned crop, separately or in mixed inoculums. The two challenged pathogenic fungi used as infested agents were Pythium ultimum and Rhizoctonia solani. The two pathogenic fungi were tested alone or in presence of the antagonizing bacterial strains each alone or mixed together.
Colonization ability of the isolate establishes a close relationship with the plant host than rhizosphere and helps in augmentation of the defense response to cope with extreme conditions. Effective colonizing bacterium encounters a protective environment where they could have a better survival and therefore more prolonged activity [30]. The bacterial strains were individually tested for in vitro antibiosis against all other. No antibiosis was observed among isolates of each mixture. Also, these isolates were tested for colonizing activity with the indicator plants and the test indicated that all isolates were able to colonize roots.
Generally, the supplementation of snap bean plants with different bacterial strains (B. thioparans and S. maltophilia) increased nitrogen contents and plant dry weight significantly but was slightly less than what obtained in the mixed inoculum treatment (Mix). Albino et al. obtained similar trends with some putative N2-fixers belonging to the genera Bacillus, Methylobacterium, Paenibacillus, Pseudomonas, and Sphangomonas isolated from the rhizosphere of some plants [31]. Similar results were obtained by Bin et al., Asghar et al., Gray & Smith and Figueiredo et al. [32-35].
Explaining the defense mechanisms against pathogenic fungi, Chen et al. stated that root and crown rot cucumber caused by Pythium can be suppressed by P. corrugate strains and the PO and PPO activities were increased in roots 2-5 days after bacterization with P. corrugate [36]. While Ramamoorthy et al. stated that P. fluorescence was found to protect tomato plants from wilt disease caused by F. oxysporium. Induction of defense protein and chemicals against challenge inoculation with the fungi in tomato plants was pronounced [26]. Phenolics were found to accumulate in bacterized tomato root tissues challenged with the fungi at one day after pathogen challenge. Activities of PO and PPO increased in bacterized root challenged with pathogen. The bacterial antagonist was found by Tendulkar et al. exhibited highest antifungal activity against the rice plant fungus M. grisea which was isolated from soil and identified as B. licheniforms BC 98 [37]. Besides M. grisea, the isolate also inhibited the growth of other phytopathogens such as Curvularia luneta and Rhizoctonia bataticola. The active material was identified as peptide surfactin. The antagonist inhibits germination of M. grisea, a potent rice phytopatogens, and therefore appears to be a potential candidate for control rice blast disease. More recently, Datta et al. stated that PGPR can enhance the growth and the productivity by exerting beneficial effects (direct or indirect) [38].
The results confirmed that combined popular as biofertilizer and bioenhancer the associations among the rhizobacterial inoculants can play a critical role in alleviating inhibitory production, providing nutrients and in stimulating each other via physical or biochemical activities that may increase the beneficial attributes of their physiology [39].
In conclusion, strains (B. thioparans and S. maltophilia) were used each alone or in combination as plant growth promoting and controlling the damping-off diseases caused by Py. ultimum and R. solani on snap bean. Addition of 1/3 recommended dose of N in presence of P and K at their recommended dose inoculated with any of antagonizer bacteria especially when they added together increase the growth and nitrogen content of snap bean plants in extreme or normal conditions.
References
- Avis TJ, Gravel V, Antoun H, Tweddell RJ (2008) Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol Biochem 40: 1733-1740.
- Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, et al. (2015) Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol Fertil Soils 51: 403-415.
- Lagos ML, Maruyama F, Nannipieri P, Mora ML, Ogram A, et al. (2015) Current overview on the study of bacteria in the rhizosphere by modern molecular techniques: a mini‒review. J Soil Sci Plant Nutr 15: 504-523.
- Singh RP, Jha PN (2017) The PGPR Stenotrophomonas maltophilia SBP-9 Augments Resistance against Biotic and Abiotic Stress in Wheat Plants. Front Microbiol 8: 1945.
- Nakkeeran S, Kavitha K, Chandrasekar G, Renukadevi P and Fernando WGD (2006) Induction of plant defence compounds by Pseudomonas chlororaphis PA23 and Bacillus subtilis BSCBE4 in controlling damping-off of hot pepper caused by Pythium aphanidermatum. Biocontrol Sci Technol 16: 403-416.
- Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, et al. (2009) Versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7: 514-525.
- Berg G, Egamberdieva D, Lugtenberg, B, Hagemann M (2010) Symbiotic Plant-Microbe interactions: Stress protection, plant growth promotion, and biocontrol by Stenotrophomonas. Symbios Stress Pp: 445-460.
- Abdelaziz S, Elbanna K, Elshahawy R (2016) Prevalence and Characterization of Putative Oligotrophic Bacteria in Fayoum Soils, Egypt. Front Environ Microbiol 2: 6-11.
- Rodriguez H, Gonzalez T, Goire I, Bashan Y (2004) Gluconic acid production and phosphate solubilization by the plant growth promoting bacterium Azospirillum spp. Naturwisseneschaften 91: 552-555.
- Saravanan VS, Subramoniam SR, Raj SA (2003) Assessing in vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Braz J Microbiol 35: 121-125.
- Loper JE, Schroth MN (1986) Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathol 76: 386-389.
- Meyer JM, Abdallah MA (1978) The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol 107: 319-328.
- Pallai R (2005) Effect of plant growth-promoting rhizobacteria on canola (Brassica napus. L.) and lentil (Lens culinaris. Medik) plants. Appl Microbiol Food Sci University of Saskatchewan, Saskatoon, Canada.
- Andro T, Chambost JP, Kotoujansky A, Cattaneo J, Bertheau Y, et al. (1984) Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J Bacteriol 160: 1199-1203.
- Renwick A, Campbell R, Coe S (1991) Assessment of in vivo screening systems for potential biocontrol agents of Gaeumannomyces graminis. Plant Pathol 40: 524-532.
- Cattelan AJ, Hartel PG, Fuhrmann JJ (1999) Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Amer J 63: 1670-1680.
- Koch E (1997) Screening of rhizobacteria for antagonistic activity against Pythium ultimum on cucumber and kale. J Plant Dis Prot 104: 353-361.
- Moore E, Arnscheidt A, Kruger A, Strompl C, Mau M )2004)Simplified protocols for the preparation of genomic DNA from bacterial cultures. Mol Microbial Ecol Manual 101: 3-18.
- Eden PA, Schmidt TM, Blakemore RP, Pace NR (1991) Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol 41: 324-325.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Kataria HR, Wilmsmeier RB, Buchenauer H (1997) Efficacy of resistance inducers, free-radical scavengers and antagonistic strain of Pseudomonas fluorescens for control of Rhizoctonia solani AG-4 in bean and cucumber. Plant pathol 46: 897-909.
- Alström S (1987) Factors associated with detrimental effects of rhizobacteria on plant growth. Plant Soil 102: 3-9.
- Ahmed S, Iqbal A, Rasool SA (2003) Bacteriocin-like inhibitory substances (BLIS) from indigenous clinical streptococci: Screening, activity spectrum and biochemical characterization. Pak J Bot 35: 499-506.
- Abdul-Baki AA, Anderson JD (1973) Vigour determination in soya bean seed by multiple criteria. Crop Sci 13: 630-633.
- Donald AH, Robert OM (1998) Determination of Total Nitrogen in Plant Tissue: Handbook of references methods for plant analysis. Pp: 75-84.
- Ramamoorthy V, Raguchander T, Samiyappan R (2002) Induction of defense-related proteins in tomato roots treated with Pseudomonas fluorescens Pf1 and Fusarium oxysporum f. sp. Lycopersici. Plant Soil 239: 55-68.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci 108: 4516-4522.
- Bosshard PP, Abels S, Zbinden R, Böttger EC, Altwegg M (2003) Ribosomal DNA Sequencing for Identification of Aerobic Gram-Positive Rods in the Clinical Laboratory (an 18-Month Evaluation). J Clin Microbiol 41: 4134-4140.
- Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD (2011) Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol 77: 3846-3852.
- Hardoim PR, Van Overbeek LS, Van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol16: 463-471.
- Albino U, Saridakis DP, Ferreira MC, Hungria M, Vinusea P, et al. (2006) High diversity of diazotrophic bacteria associated with the carnivorous plant Drosera villosa var. villosa growing in oligotrophic habitats in Brazil. Plant Soil 287: 199-207.
- Bin L, Smith DL, Ping-Qui F (2000) Application and mechanism of silicate bacteria in agriculture and industry. Guizhou Sci 18: 43-53.
- Asghar HN, Zahir ZA, Arshad M, Khalig A (2002) Plant growth regulating substances in the rhizosphere: Microbial production and functions. Adv Agron 62: 146-151.
- Gray EJ, Smith DL (2005) Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol Biochem 37: 395-412.
- Figueiredo MVB, Burity HA. Martinez CR, Chanway CP (2008) Alleviation of drought stress effects in common bean (Phaseolus vulagaris) by co-inoculation Paenibacillus polymyxa and Rhizobium trpici. Applied Soil Ecol 40: 182-188.
- Chen C, Bélanger RR, Benhamou N, Paulitz TC (2000) Defence enzymes Induced in cucumber roots by treatment with plant growth promoting rhizobacteria (PGPR) and Pythium aphanidermatum. Physiol Mol Plant Pathol 56: 13-23.
- Tendulkar SR, Saikumari YK, Patel V, Raghotama S, Munshi TK, et al. (2007) Isolation, purification and characterization of an antifungal molecule produced by Bacillus licheniformis BC98, and its effect on phytopathogen Magnaporthe grisea. J Appl Microbiol 103: 2331-2339.
- Datta M, Palit R, Sengupta C, Pandit MK, Banerjee S (2011) Plant growth promoting rhizobacteria enhance growth and yield of chilli (Capsicum annuum L.) under field conditions. Aust J Crop Sci 5: 531-536.
- Bashan Y (1998) Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol Adv 16: 729-770.
Copyright: © 2018 Abdelaziz S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.