
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2024)Volume 7, Issue 2
The treatment of Glioblastoma Multiforme (GBM), a highly malignant brain tumor, is critically hindered by the ineffectiveness of current modalities such as surgery, radiation and chemotherapy. These traditional methods fail to completely remove the tumor mass and lack the ability to discriminate between cancerous and normal brain cells, often resulting in collateral damage to healthy tissue and recurrence of the disease. This underscores an urgent necessity to develop novel therapeutic strategies that can target tumor cells with precision, offering hope for improved survival rates and quality of life for GBM patients. This study investigates targeted therapy, focusing on the Integrated Stress Response (ISR) that cancer cells harness to survive hypoxic stress. Specifically, it demonstrates that Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1 (EIF2AK1), which encodes Heme-Regulated Inhibitor (HRI) kinase, is activated under hypoxia and co-expressed with the glioma stem cell marker Sex Determining Region Y-box 2 (SOX2), which specifically happens in glioma cells, increasing the targeted accuracy of the repurposing drug. This correlation, indicating hypoxia-driven stemness, is confirmed at both the genetic level and through Gene Set Enrichment Analysis (GSEA). Furthermore, GSEA in spatial transcriptomics shows hypoxia-induced glycolysis, disrupting the tumor microenvironment and causing necrotic cell death. Stemness phenotype is induced in the peripheral cells due to the unfavourable hypoxic environment. Hemin, an HRI inhibitor, has been repurposed to inhibit ISR and mitigate hypoxia. Treatment with hemin on the Uppsala 87 Malignant Glioma (U87 MG) cell line resulted in IC50 values of 23.50 μM and 52.46 μM at 24 and 48 hours, respectively, surpassing Temozolomide's efficacy. A decrease in HRI expression after the hemin treatment suggests the and ISR activity and, potentially, hypoxia. This would reverse the unfavourable microenvironment so that the stemness phenotype doesn’t spread. Potentially, invasiveness and recurrences of GBM in clinic situation would decrease, thus potentially improving patient prognosis. The therapeutic potential of hemin is enhanced by its ability to kill glioma cells directly and accurately in the glioma cell in original Tumor Microenvironment (TME) when cells are proliferating with adequate oxygen. Therefore, this study demonstrates the therapeutic potential of repurposing hemin, an HRI inhibitor, to precisely target hypoxia-induced glioma stem cells in glioblastomas, disrupting the aggressive TME to potentially improve patient prognosis.
Glioma; Stemness; Targeted therapy; Integrated stress response; Multi-omics analysis
Gliomas encompass a broad spectrum of tumors originating from glial cells in the brain and spinal cord. Treating this diverse group presents significant challenges due to their unique anatomical locations and the increasing difficulty of treatment as glioma grade escalates. Gliomas are classified based on cell type, grade and location, with grades I and II referred to as Low-Grade Gliomas (LGGs) and grades III and IV as High-Grade Gliomas (HGGs). Among these, Glioblastoma Multiforme (GBM) is the most aggressive form and the primary focus of this study. GBM is notorious for its dismal prognosis, with a median survival time of just 12-18 month’s post-diagnosis. Despite advancements in medical treatments, the five-year survival rate remains around 5%-10%. This bleak outcome is due to the tumor's aggressive behavior, high recurrence rate and resistance to standard therapies. Diagnosis typically involves neurological examination, imaging tests such as Magnetic Resonance Imaging (MRI) and often a biopsy to determine the tumor's type and grade. Treatment usually consists of a combination of surgery, radiation therapy and chemotherapy, with the extent of surgical resection being a critical prognostic factor. However, the specific location of the glioblastoma often limits surgical options. Post-surgery radiation therapy combined with chemotherapy targets residual tumor cells [1-3].
Nevertheless, standard treatment modalities face significant limitations. Surgical interventions in glioma treatment may not ensure complete tumor removal due to the tumor's infiltrative nature and proximity to critical brain structures, leading to a high risk of recurrence. While radiation therapy can shrink tumors and control growth, it may also damage surrounding healthy brain tissue, causing long-term neurological side effects. Additionally, its efficacy against cancer stem cells, which are often more resistant to radiation, is limited, potentially contributing to tumor recurrence and progression. Chemotherapy, meanwhile, can cause non-specific toxicity to healthy cells, resulting in side effects such as nausea, fatigue and immunosuppression. Moreover, glioma cells, especially cancer stem cells, may develop resistance to chemotherapeutic agents. Given these challenges, there is a pressing need for novel therapeutic approaches [4].
Targeted therapy offers several advantages over traditional treatment methods by inhibiting specific molecular targets associated with cancer growth and progression. This approach allows for more precise and effective treatments with potentially fewer side effects [5].
This study focuses on the Integrated Stress Response (ISR) in GBM, a cellular adaptation mechanism to various stressors, such as hypoxia or nutrient deprivation. The ISR plays a crucial role in tumor survival, growth and therapy resistance, making it a promising target for GBM treatment strategies. Specifically, this research examines the role of hypoxia in the ISR pathway, highlighting the transcription and activation of EIF2AK1, encoding the HRI, leading to the phosphorylation of eukaryotic translation Initiation Factor 2 Alpha (eIF2α) [6,7]. This phosphorylation, regulated by activated sensor kinases, reduces global protein synthesis while selectively enhancing the translation of certain messenger Ribonucleic Acid (mRNAs), such as Activating Transcription Factor 4 (ATF4), which activates an adaptive response aiding glioma cells to become more malignant. In GBM, the ISR enhances the tumor's malignancy by improving its survival, invasiveness and treatment resistance, particularly under hypoxic conditions. Inhibiting HRI with agents like hemin disrupts the adaptive stress response, potentially impairing the tumor's ability to cope with environmental stressors via the ISR pathway and reducing its survival and growth prospects. Nonetheless, the stress would still be managed with the iron molecule in hemin, which increases the oxygen-carrying capacity of the TME, thereby relieving the hypoxic condition.
Transcriptomics analysis, particularly single-cell Ribonucleic Acid sequencing (scRNA-seq), is employed to identify EIF2AK1 and its co-expression with the stem cell marker SOX2. This technique allows for gene expression analysis at the single-cell level, revealing cellular heterogeneity within tumors and enabling the identification of specific cell types and states within complex tissues. Spatial transcriptomics further enhances this analysis by mapping gene expression to specific tissue locations, offering valuable insights into the tumor microenvironment and cancer progression [8].
This study aims to repurpose hemin as a novel therapeutic agent targeting one of the initial kinases in the ISR pathway. An analysis of scRNA-seq and spatial transcriptomics data shows a strong co-expression and correlation between EIF2AK1 and SOX2, underscoring the link between ISR and glioma stemness. Subsequently, the efficacy of EIF2AK1 is observed through its inhibition by hemin in the cell viability assay. The expectation was that inhibition by hemin would lead to a decrease in the expressions of EIF2AK1 and SOX2, as confirmed by western blot analysis. While other pathways may also contribute to inhibiting cancer cell growth, the ISR pathway represents a critical mechanism for curbing the proliferation of cancer cells.
Dataset collection and preprocessing
The single-cell RNA-sequencing (scRNA-seq) dataset (GBM_ GSE117891_10X) was downloaded from the GEO with only the GBM patients [9]. The spatial transcriptomics RNA-sequencing (stRNA-seq) dataset was downloaded from the 10 × Genomics datasets [10]. Bulk RNA-seq dataset is downloaded from the Ivy Gap dataset, with profiling information within different histological and verhaak cell types [11]. The corresponding clinical data from the TCGA database portrays the patient survival plots [12].
Single-cell sequencing and spatial transcriptomics data processing
I used the R package Seurat (v4.1.0) to process single-cell and spatial transcriptomics data. I first used the “read.delim” function to read the cell matrix profile downloaded from the Gene Expression Omnibus (GEO) website. Then, the function “Create Seurat Object” was applied to convert the matrix into a Seurat object. I excluded those cells with fewer than 200 genes and more than 10000 genes. I log-normalized the Seurat object and identified highly variable features using the “Find Variable Features” function with the parameter’s selection (method=vst and nfeatures=2500). I subsequently scaled the Seurat object and performed linear dimensional reduction using the Run Principal Component Analysis function with the variable features of the Seurat object. I visualized the distribution of each principal component using the “Elbow Plot” function and used the first 15 principal components for clustering. I performed K-nearest neighbors clustering for the Seurat object with the parameter dims=1:25 through “Find Neighbors” function. I performed Find Clusters function with the parameter resolution=0.3 and Uniform Manifold Approximation and Projection (UMAP) clustering using the Run UMAP function with the parameter dims=1:25. I then identified the cell type of each cell cluster according to the differentially expressed gene for each clusters. For spatial transcriptomics of GBM, I applied the function “SC Transform” to normalize the data of spatial transcriptomics. I used functions “Run PCA”, “Find Neighbors” and “Find Clusters” to reduce the dimensionality and cluster similar spatial spots [13-15].
Trajectory analysis
First, from the preliminary scRNA-seq analysis, cell types related to cancer cells are isolated. Within those cancer cells, the starting point of the trajectory analysis is stage 2 GBM patients with a preliminary form of the cancer cell type. I learned trajectory graphs and analyzed pseudo time using the monocle 3 (learntrajectory) function [16].
Gene set enrichment analysis
To investigate differences in transcriptome and function between cell types in scRNA-seq and stRNA-seq data, I used the function “Find All Markers” of Seurat to find Differentially Expressed Genes (DEGs) of cell types in the GBM, DEGs of each cell type were used for visualization. Then, Gene Set Enrichment Analysis (GSEA) of DEGs was performed to determine the enrichment score of oncogenic hallmark pathways in malignant cells (p-value<0.05). Differentially expressed genes were also applied in different malignant subtypes. The oncogenic hallmark pathways gene sets (h.all.v7.1.symbols) were downloaded from the Molecular Signatures Database (MSigDB) database [17].
In addition, the GSEA analysis on spatial transcriptomics is done on Spatial Transcriptomics Analysis Tool (SPATA2), a cutting-edge method that combines histological imaging and genomic analysis. It allows researchers to see and measure gene expression in tissue sections in detail. It stands out because it gives you a single platform for processing and seeing this complicated data. It makes finding gene expression patterns in different areas easier, which lets users connect these patterns to specific tissue shapes. SPATA2 is a powerful tool for researchers who want to get useful information from spatial transcriptomics datasets because it has an easy-to-use interface and powerful analytical features.
Patient survival
The TCGA database is used to determine patient survival. Specifically, the patient survival was compared between high expression and low expression of EIF2AK1, where the maximally selected rank statistics determines the threshold and the results of the comparison were summarized in a Kaplan Meier survival curve with the corresponding p-values attached [18]. The Kaplan-Meier survival curve figures out how likely it is that someone will live over time and determines the chances of survival at each time point that has been observed, like when someone dies and then plots these odds on a graph. Considering censored observations, the curve shows what percentage of subjects will survive over a certain period.
Cell culture and cell viability test
Cultured cells are grown in flasks under standard cell culture conditions with Dulbecco’s Modified Eagle’s Medium (DMEM). The cell cultures are incubated at 37°C and 5% CO2 and the cells are subcultured after each 24 hours. Following a 48-hour incubation period, which allows for sufficient cell growth and attachment, these cells are carefully transferred to a 96-well plate for drug treatment. Different concentrations of the drug (hemin and temozolomide) are added to the respective wells of the plate. This setup includes control wells where no drug is added to serve as a baseline for comparison. The cell viability is assessed at two time points post-drug treatment: at 24 hours and again at 48 hours. Each concentration at a time point is repeated three times. Cell Titer-Glo is employed as the detection method for cell viability by measuring the ATP content for this assessment [19].
Western blotting
Western blotting was performed on cells from the U87 cell line. Proteins were extracted in Radio-Immunoprecipitation Assay (RIPA) lysis buffer containing protease and phosphatase inhibitor cocktails, separated by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and transferred onto Polyvinylidene Fluoride (PVDF) membranes. 5% bovine serum albumin in Tris Buffer (TBS) was used for membrane blocking overnight at 4°C and the membrane was then washed with Phosphate-Buffered Saline (PBS) containing 0.1 % Tween 20. The antibodies were against SOX2 and EIF2AK1, with catalog numbers AF2018 and 67674-1-Ig, respectively.
Primary antibodies specific to target proteins were used for probing for 1 h at room temperature and then washed with Tris-Buffered Saline with 0.1% tween 20 (TBST) again; corresponding secondary antibodies were used for detection [20].
Profiling of glioblastoma scRNA-seq data and identification of cell type associate with cancer stemness
This study started by analyzing the scRNA-seq data of 12 glioma patients from grade II to grade IV, which gave a basic understanding of the dataset, including the differentially expressed gene, cell type information and cell lineage within the cancer subset. Initially, the cells were clustered into 9 different cell types, including immune cells, mesenchymal cells, oligodendrocytes and cancer cells labeled based on the verhaak subtypes [21-22].
UMAP visualization by patients showed all clusters were contributed by all patients, except cluster 5 and 7, which only consisted of cells from patient 1. This also suggested the minimization of batch effects [23]. I further shown the UMAP visualization of all different clinical stages, including high-grade gliomas, like GBM or low-grade glioma. A cluster of pro-neural cluster merely derived from the grade II patients, indicating the stemness of this cluster.
The normalized expression of nine most prominent marker genes was shown on the feature plot. Fork head box protein J1 (FOXJ1) specifically indicated the ependymal subtype derived from patient 13. Also, some very distinct marker genes appear, such as Myelin-Associated Glycoprotein (MAG), which stood for the general category of oligodendrocytes. To investigate the cell type more closely, marker genes of each cluster were specifically studied.
In total, 20 clusters were identified with a 0.3 resolution Louvain clustering. Among those 20 clusters, 9 of them were identified as GBM cancer cell clusters marked by the expression of SOX2 and EGFR. Within these clusters, clusters 0, 5 and 7 had high expression of Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A), which were labeled as CDKN2A malignant 1-3. Cluster 7 also specifically expressed FOXJ1, indicating ependymal cell features.
In clusters 6 and 12, the high expression of Platelet-Derived Growth Factor Receptor Alpha (PDGFRA) indicated that these two clusters were pro-neural subtypes under the Verhaak categories. Cells in cluster 2 expressed high levels of cell cycle genes such as Topoisomerase 2 Alpha (TOP2A), Marker of Proliferation (MKI-67), indicating active proliferation and high malignancy. Cluster 14 and 17 were identified as the classical glioma cell clusters marked by the high transcript levels of Epidermal Growth Factor Receptor (EGFR). They represented the most typical glioma cell expression in the solid tumor and they lack the special characteristics of stemness and malignancy and thus are easier to treat.
In addition, I also identified various immune cell subtypes in the glioma microenvironment, which primarily include macrophage/ microglia, monocytes and T cells. T cells (cluster 11), marked by the expression of CD3G, had low abundance in the tumor microenvironment compared to the diverse innate immune cells, which were defined by the high expression of Colony Stimulating Factor 1 Receptor (CSF1R). Within those innate immune cells, also shown in Figure 1, two of the largest clusters, clusters 1 and 8, were identified as the macrophages/monocytes marked by the high expression of Lysosomal-Associated Protein Transmembrane 5 (LAPTM5) and Cluster of Differentiation 14 (CD14) [24]. These genes were responsible for activating macrophages and signal transduction, respectively.
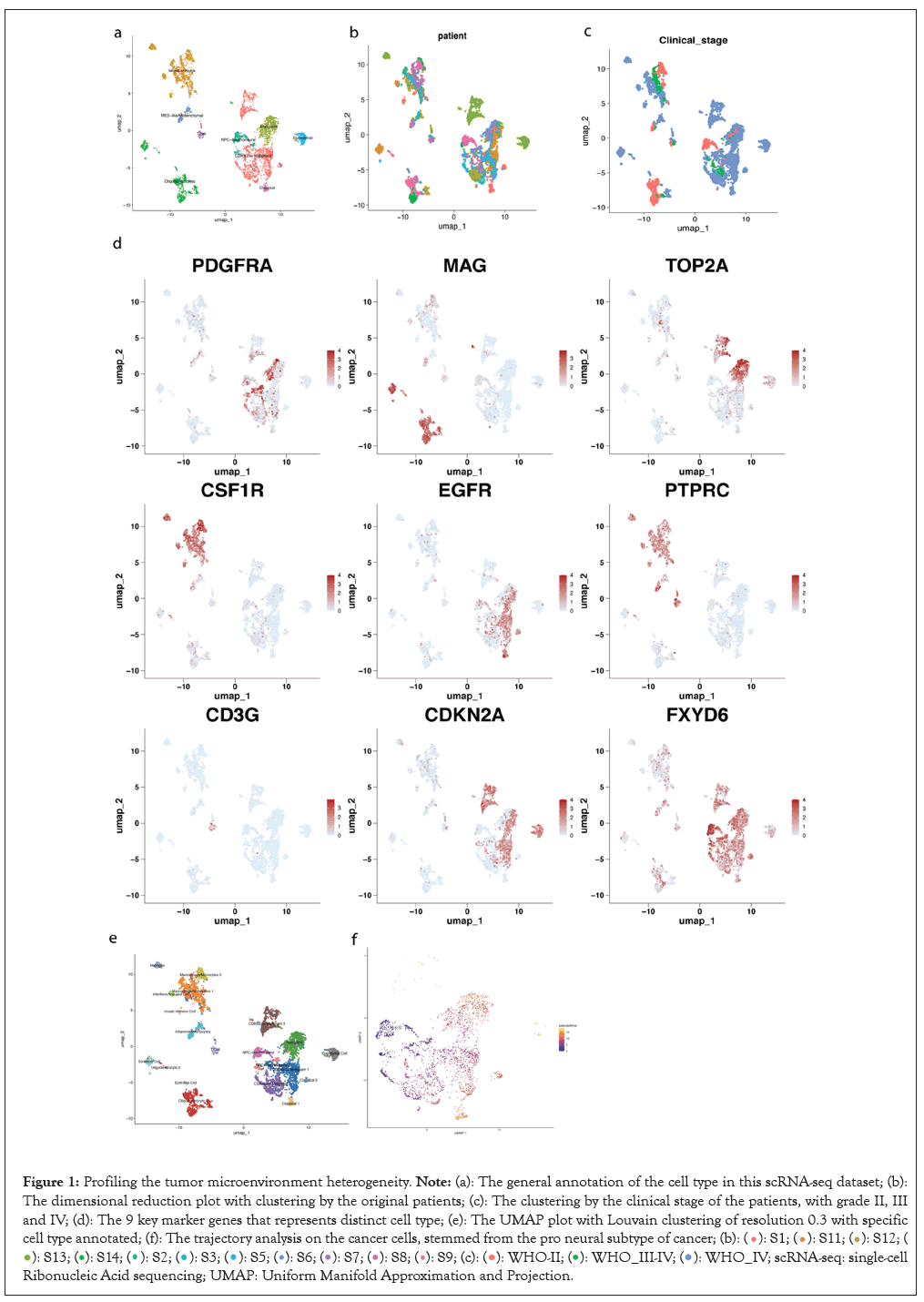
Figure 1: Profiling the tumor microenvironment heterogeneity. Note: (a): The general annotation of the cell type in this scRNA-seq dataset; (b):
The dimensional reduction plot with clustering by the original patients; (c): The clustering by the clinical stage of the patients, with grade II, III
and IV; (d): The 9 key marker genes that represents distinct cell type; (e): The UMAP plot with Louvain clustering of resolution 0.3 with specific
cell type annotated; (f): The trajectory analysis on the cancer cells, stemmed from the pro neural subtype of cancer; 
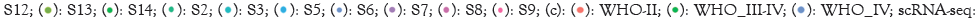 single-cell Ribonucleic Acid sequencing; UMAP: Uniform Manifold Approximation and Projection.
single-cell Ribonucleic Acid sequencing; UMAP: Uniform Manifold Approximation and Projection.
In addition, cluster 15 was identified as the interferon-induced cells since they highly expressed SIGLEC1, which was reported to increase in circulating monocytes when induced by type I interferon [25]. Similarly, cluster 9 was identified as the inflamed monocytes due to the high expression of genes that encoded interferons and corresponding receptors, such as Interleukin-1-Beta (IL1B) and Interleukin 1 Receptor type 2 (IL1R2).
Cluster 13 was identified as the microglia for its high production of P2RY12, which is a key marker for identifying homeostatic microglia, as opposed to inflamed microglia [26]. Cluster 16 was simply labeled as an innate immune cell since it had a small number of cells with little distinctive information.
Also, the distribution of that cluster was not centered together since the right part of this cluster is close to cluster 1, which was the macrophage/monocyte cluster and the left part was more in between the interferon clusters. This suggested a potential mixture of cell status or doublet contamination, contributing to the difficulties in identifying a clear cell subtype.
The last piece of this UMAP plot was centered around the bottom left corner, mostly oligodendrocyte-related cell types by the high expression of the general oligodendrocyte marker MAG.
Cluster 3, as the largest cluster in this region, took the identity of oligodendrocytes since it had a high expression in typical markers like Oligodendrocyte Transcription Factor 1 (OLIG1) and Oligodendrocyte Transcription Factor 2 (OLIG2) and it also had higher cell counts than cluster 10, which would be logical since oligodendrocytes are the predominant cell type in this larger cluster identified by the MAG since GBM is a Central Nervous System (CNS) related disease and oligodendrocytes occurs more in CNS [27].
In addition, cluster 10 was identified as the Schwann cell for its high expression in S100 series genes, which is useful for identifying neoplasms derived from the Schwann cells. These genes included S100A1, S100B, S100A13. In addition, SOX10, which is more specific to the Schwann cells, is also highly expressed in cluster 10 [28].
Clusters 18 and 19 were identified as oligodendrocytes and epithelial cells, respectively. Cluster 18 had a similar gene expression profile compared to cluster 3 with strong oligodendrocyte characteristics and cluster 19 had key marker genes like Gremlin-1 (GREM1) and Troponin C1 (TNNC1), which are responsible for the generation of connective tissue between cells and thus are classified as epithelial cells.
To examine the potential differentiation trajectory of various subtypes of cancer cells, pseudotime analysis was performed via monocle. These two clusters, clusters 6 and 12, identified as pro-neural subtypes with strong stemness features, were set as the root state.
Cluster 0, 4 and 14 were found at an intermediate differentiation stage. In contrast, clusters 2, 5 and 7 were shown as the most differentiated cancer cells. It is important to learn about the cancer progression and lineage since the stemness of cancer cells drives not only proliferation but also resistance to drugs and apoptosis (Figure 1) [29-31].
Hypoxic gene EIF2AK1 as a key component associated with stemness gene SOX2
To identify what biological pathways were highly enriched for the glioma cancer cells, especially the cells with stemness, I performed GSEA analysis and found they were enriched for the stemness pathways and hypoxia pathway. Integrative Stress Response (ISR) pathways were well known to be induced and promote cell survival during a hypoxia environment.
Therefore, I performed correlation analysis to examine the co-expression of ISR genes with SOX2, one of the most typical stemness markers in the Ivy Glioblastoma Atlas dataset. I found EIF2AK1, one of the initiators for the ISR pathway, has strong positive co-expression with SOX2.
Analyzing the expression patterns of EIF2AK1 and SOX2 across different pathological locations in the tumor showed that other than the regular cellular tumor and infiltrating tumor, which would be expected to have the most expression due to their direct location and relation to the tumor, the pseudopalisading cells were also enriched in both gene expression, indicating that both gene was specifically also enriched in a hypoxic TME [32].
The co-expression on the single cell level was then further confirmed by demonstrating high expression of both genes shown in white dots.
Other than directly visualizing the co-expression location and level with the dots and the color scale, the co-expression also distributed across multiple cell types. Immune cells had the least amount of co-expression, meanwhile the oligodendrocytes and Schwann cells also showed a low amount of co-expression because they were myelinating cells conducting signals between neurons with low metabolic levels after they are myelinated [33].
The results above indicated that cancer cells were the major cell type consuming oxygen since they possessed many stem-like characteristics. As a result, this co-expression was relatively specific to the cancer part of the brain and thus provided potential for targeted therapy.
To determine the potential prognostic values of EIF2AK1, a 13 genes signature that composed the EIF2AK1 signaling pathway were used to determine enrichment scores in glioma specimens based on data from The Cancer Genome Atlas (TCGA). Enrichment of the EIF2AK1 signature was associated with worse patient overall survival.
This survival analysis gave insights for the in vitro experiments to test out the therapeutic effects since inhibition of EIF2AK1 was a reasonable choice due to longer survival time with lower gene expression. Since hypoxia induces stemness, the inhibition of EIF2AK1 by relieving the stress would, expectedly, should produce a lower SOX2 expression (Figure 2) [34].
Figure 2: The co-expression of EIF2AK1 and SOX2 and associations with prognosis. Note: (a): The GSEA data of differential expressed genes from
cancer subtypes in winter hypoxia pathway; (b): The Gene Set Enrichment Analysis (GSEA) data of differential expressed genes from cancer
subtypes in ramalho stemness pathway; (c): The correlation between EIF2AK1 and SOX2 in expression level; (d): The EIF2AK1 mRNA expression
level categorized by different histology regions; (e): The SOX2 messenger Ribonucleic Acid (mRNA) expression level categorized by different
histology regions; (f): The expression of Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1 (EIF2AK1) and Sex Determining Region Y-box
2 (SOX2), respectively and the coexpression plot with a scale on the right most figure; (g): The survival plot between high and low expression of EIF2AK1 signature genes in the TCGA dataset;  EIF2AK1_sig2_Lo.
EIF2AK1_sig2_Lo.
Hemin efficiently kills glioblastoma cells and reduces the protein expression levels of EIF2AK1
To explore whether inhibiting EIF2AK1 could be a novel therapeutic strategy for glioma patients by suppressing the expression of SOX2, I designed experiments to test the treatment efficacy of hemin-a EIF2AK1 inhibitor in vitro. The choice of hemin as the inhibitor of HRI (encoded by EIF2AK1) is because it is an FDA-approved drug and it is also directly related to the function of HRI, which is produced with iron-deficiency and hemin exactly provides iron to remediate this problem [35-37]. Temozolomide served as a positive control, which had already been on the market for treating GBM. The data from the temozolomide group matched with past literature, so the data from the hemin group was more convincing [38-40]. The choice of the U87 cell line was valid since the cell line expresses SOX2 and EIF2AK1, so the ISR pathway and related pathways should be involved in this cell line [40-41].
The cell culture and cell viability experiment were used to determine the effect of hemin on killing glioma cells. In vitro, the cell viability test showed that, in the U87 cell line, hemin had a great sensitivity to hemin. The IC50 was determined by a logistics regression with the 3 data points for each drug concentration, shown in the error bar. For 24 h and 48 h hemin treatment, the IC50 was 23.50 μM and 52.46 μM respectively. The explanation for this was that the glioma cell proliferates quickly, so it would take less drug when the drug only proliferates for 24 hours. After another 24 hours, it would need more than double the amount to kill half of the glioma cells. Compared to the control group with Temozolomide with an IC50 of 156.7 μM, hemin showed a lower IC50 than Temozolomide, suggesting that it was more effective at killing cancer cells. With proper drug delivery technology, hemin would work best if they can accurately target cancer cells.
To test if hemin kills cancer cells by lowering the expression of EIF2AK1 and SOX2, I performed western blot analysis to compare the protein expression of EIF2AK1 and SOX2 with or without hemin treatment. The western blot showed that both HRI and SOX2 decrease after a 48-hour hemin treatment, compared to the control protein Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH). This showed that the ISR pathway for hypoxia is reduced in terms of activity. HRI would activate EIF2A in ISR and with that decrement for HRI, EIF2A would also be less phosphorylated, thus decreasing the activity of the pathway (Figure 3).
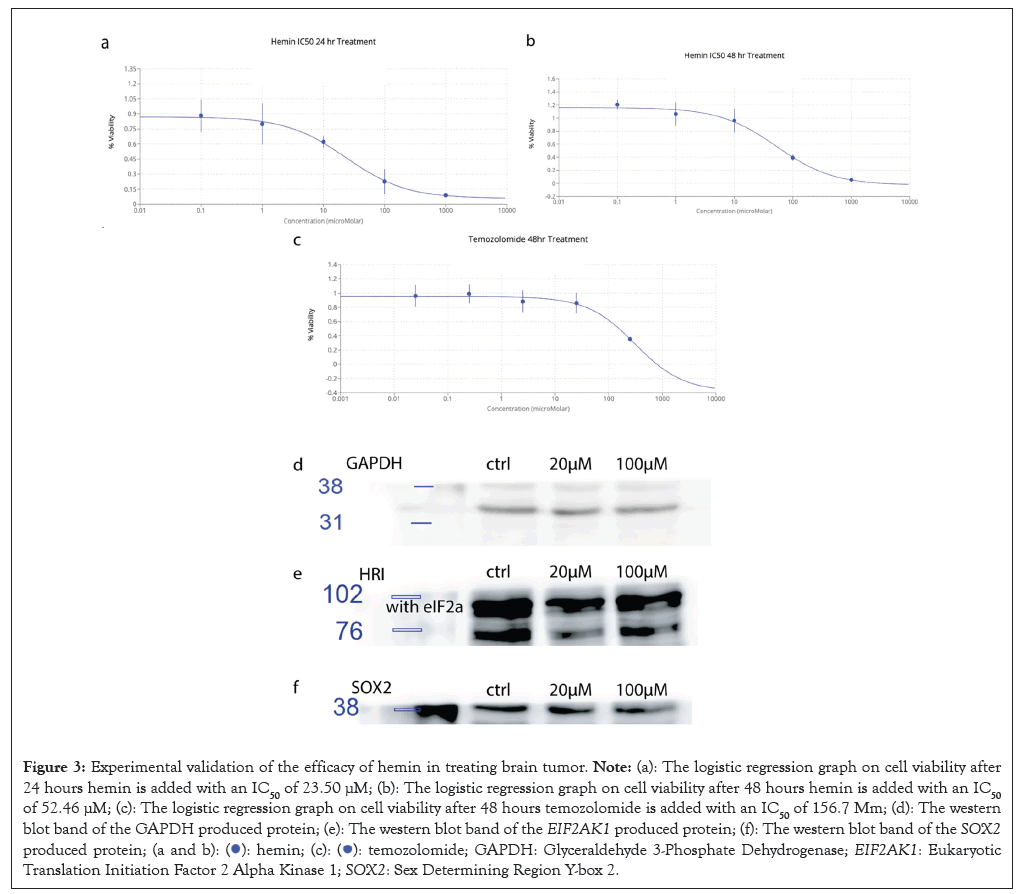
Figure 3: Experimental validation of the efficacy of hemin in treating brain tumor. Note: (a): The logistic regression graph on cell viability after 24
hours hemin is added with an IC50 of 23.50 μM; (b): The logistic regression graph on cell viability after 48 hours hemin is added with an IC50 of
52.46 μM; (c): The logistic regression graph on cell viability after 48 hours temozolomide is added with an IC50 of 156.7 Μm; (d): The western
blot band of the GAPDH produced protein; (e): The western blot band of the EIF2AK1 produced protein; (f): The western blot band of the SOX2 produced protein;  GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase; EIF2AK1:
Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1; SOX2: Sex Determining Region Y-box 2.
GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase; EIF2AK1:
Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1; SOX2: Sex Determining Region Y-box 2.
Spatial transcriptomics shows similar co-expression and suggests hypoxia leads to necrosis and migration of glioma cells
In spatial transcriptomics, the pathways were specifically located on the tissues spatially so that the area that the pathways were enriched can be visualized, compared to the GSEA done on the cancer cluster in general, which cannot be used to determine whether the same cell was responsible for both hypoxia and stemness enrichment. Those co-expressions and GSEA analyses on tumor tissues enabled further exploration of TME and the spatial distribution of the enriched pathways, giving a more accurate understanding of the location of stemness, hypoxia and other pathways that might be related to those two pathways due to similar spatial distributions.
The hypoxia enrichment was uniform, though not the most significant, shown in the lower left region of the tumor slice. More evidently, the glycolysis was also enriched in the same area with both uniformity and significantly higher expression. This could be explained by saying that not enough oxygen is taken in that specific region, so the glioma cells cannot normally metabolize as they could not perform oxidative phosphorylation due to the lack of oxygen. As a result, the only way they can generate Adenosine Tri Phosphate (ATP) is through glycolysis. However, this was not a long-term solution as the lactate is produced, altering the pH of the microenvironment by attracting hydrogen. With the lack of oxygen supplied by the blood, cell necrosis also took place in that same region.
Moreover, the opposing region on the top right of this tumor slice showed great stemness and cell cycle pathway enrichment. This was caused by glioma stem cells proliferating with adequate oxygen around the hypoxia region. However, this also suggested that the hypoxia region promotes tumor invasiveness since the hypoxic region was no longer suitable for the glioma cells to grow and leads to an increase in glycolysis and necrosis, the glioma cells would choose to migrate to the TME and keep proliferating where there was enough oxygen to metabolize (Figure 4).
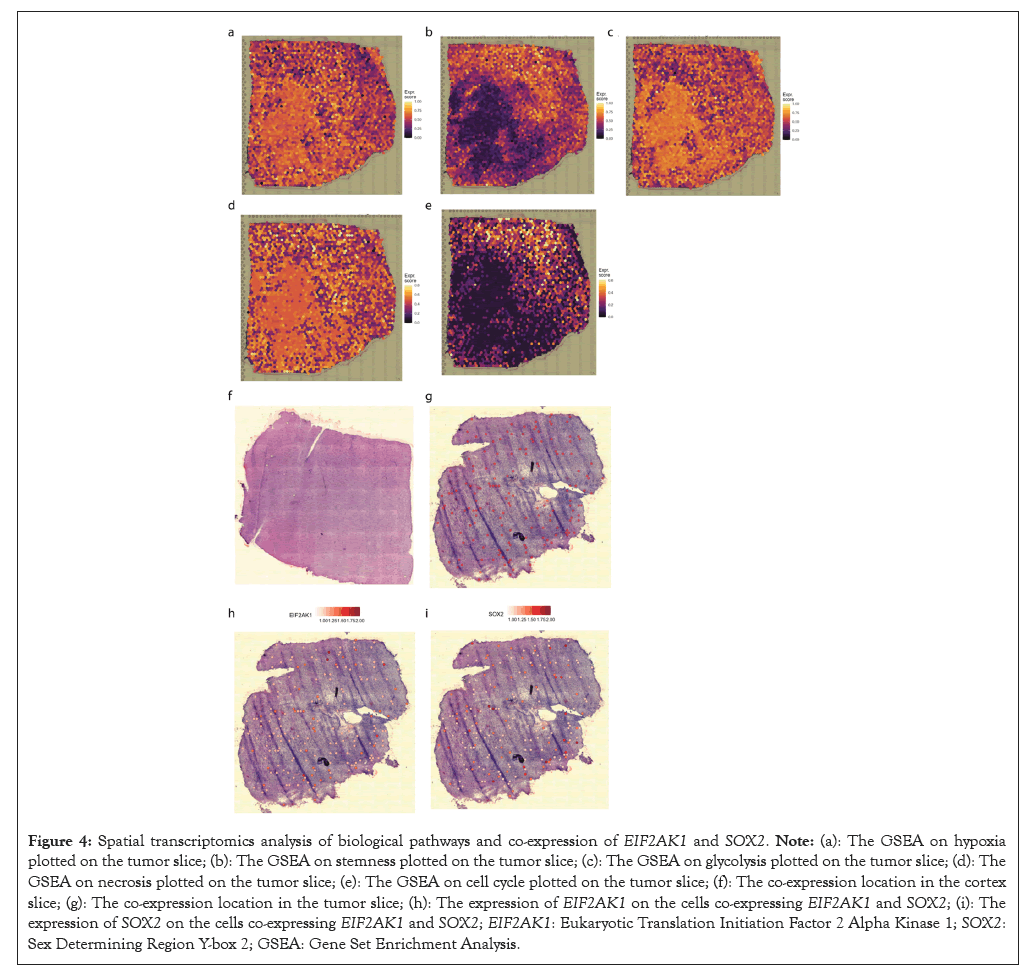
Figure 4: Spatial transcriptomics analysis of biological pathways and co-expression of EIF2AK1 and SOX2. Note: (a): The GSEA on hypoxia plotted on the tumor slice; (b): The GSEA on stemness plotted on the tumor slice; (c): The GSEA on glycolysis plotted on the tumor slice; (d): The GSEA on necrosis plotted on the tumor slice; (e): The GSEA on cell cycle plotted on the tumor slice; (f): The co-expression location in the cortex slice; (g): The co-expression location in the tumor slice; (h): The expression of EIF2AK1 on the cells co-expressing EIF2AK1 and SOX2; (i): The expression of SOX2 on the cells co-expressing EIF2AK1 and SOX2; EIF2AK1: Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1; SOX2: Sex Determining Region Y-box 2; GSEA: Gene Set Enrichment Analysis.
This study built upon existing literature that linked hypoxia to the stemness of cancer cells, applying this concept to the Integrated Stress Response (ISR) specifically. In this research, EIF2AK1 emerged as a novel target for treating hypoxia, enriching the potential treatment avenues by introducing a biological pathway. The inhibition of EIF2AK1 with hemin specifically disrupted the ISR pathway, which was observed to reduce tumor growth and preserve genomic stability by decreasing cell cycle arrest-a process that often leads to mutations due to repeated genome duplications.
Beyond the direct interruption of the ISR, the hypoxia typically induced in the tumor microenvironment was also alleviated. The iron supplied by hemin facilitated oxygen transport, as iron is a component of hemoglobin, increasing oxygen availability in the tumor microenvironment. Consequently, HRI expression diminished. This oxygen enrichment also contributed to genomic stability and lessened the likelihood of mutations, reducing the risk of normal cells transforming into cancerous ones. As a result, the adverse effects of hypoxia were mitigated through hemin administration.
The western blot results established a decrease expression in EIF2AK1 and SOX2. Although a direct relationship between EIF2AK1 and SOX2 was not established, the observed co-expression and correlation between these genes implied an indirect interaction. Hence, the reduction in EIF2AK1 expression appeared to correspond with a similar decrease in SOX2 levels. This decline in SOX2 indicated diminished stemness within glioma cells, which could slow cell proliferation and differentiation, potentially reducing cancer migration. Analysis of spatial transcriptomics data supported this, showing areas of severe hypoxia adjacent to less hypoxic regions where induced stemness was decreased. Consequently, in clinical scenarios, this reduction in stemness and migration could diminish the likelihood of tumor recurrence, potentially transforming GBM into a more manageable long-term condition and enhancing patient survival times.
Furthermore, hemin exhibited pronounced cytotoxicity in the U87 MG glioma cell line. As tumor migration was confined to specific local Tumor Microenvironments (TMEs), glioma cells tended to expand within these areas, where hemin effectively induced cell death due to its inherent toxicity. Hemin's targeting was more precise since it naturally gravitated towards hypoxic regions, leading to cell death before the cells could fully recover from hypoxia.
Similar studies also corroborate the findings in this study. Ravi, et al. [10] suggested that hypoxia can lead to cell cycle arrest, which shows decreased tumor proliferation in a specific tumor microenvironment. This did not necessarily contradict the relationship with stemness, as stemness encompasses the cell's ability to perpetuate its lineage and interact with its environment, balancing between quiescence, proliferation and regeneration [42]. This means that stemness doesn’t necessarily mean that the cell is proliferating at a high speed, especially in this hypoxic environment. Instead, they demonstrated their stemness by migrating toward TME with normal oxygen content, which is shown by the stemness and cell cycle pathway enrichment in my spatial transcriptomics analysis. Additionally, the genomic instability in hypoxic regions increased due to cell cycle arrest during the Synthesis (S) phase, leading to copy number variations and a higher mutation rate due to the duplicated chromosomes.
Hemin's efficacy in reducing glioma cell viability warrants further testing under hypoxic conditions in vitro, to better simulate the oxygen depleted TME. Further in vivo tests could provide a more accurate assessment of the drug's efficacy and toxicity within an animal model.
Future work from this study could include siRNA transfection experiments to knock down EIF2AK1 expression, clarifying the direct impact of ISR pathway inhibition on tumor suppression. Considering glioblastoma's brain localization, assessing hemin's ability to traverse the Blood-Brain Barrier (BBB) is crucial. Should hemin be unable to penetrate the BBB, alternative drug delivery methods must be explored, such as transient BBB disruption with ultrasound or chemically modifying hemin for BBB permeability [43].
In summary, current treatments for Glioblastoma Multiforme (GBM) are limited in their effectiveness, necessitating the development of precise therapeutic strategies. This study identified the co-expression of Eukaryotic Translation Initiation Factor 2 Alpha Kinase 1 (EIF2AK1) and Sex Determining Region Y-box 2 (SOX2) in cancer cells, aligning with pan-cancer research that associates hypoxia with increased cancer cell stemness. I observed significant anti-tumor effects in cell cultures by targeting the Integrated Stress Response (ISR) and repurposing hemin as a therapeutic agent against Heme-Regulated Inhibitor (HRI). The inhibition of EIF2AK1 by hemin also suggested a reduction in cancer cell stemness, promising a novel approach to glioblastoma treatment.
I would like to thank my parents for funding my project, Himanshu Dashora who helped me with the exploratory analysis, which initiates the idea of this research and Dayu Teng who helped me with the wet lab portion of the research.
Xingchuan Ma conducted the computational analysis part, including scRNA-seq and spatial transcriptomics. Xingchuan Ma and Dayu Teng conducted the in vitro and western blot.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Ma X (2024) HRI Inhibition by Hemin as a Novel Targeted Therapy for Glioblastoma via the Integrated Stress Response. J Clin Chem Lab Med. 7:286.
Received: 19-Apr-2024, Manuscript No. JCCLM-24-30863; Editor assigned: 22-Apr-2024, Pre QC No. JCCLM-24-30863 (PQ); Reviewed: 06-May-2024, QC No. JCCLM-24-30863; Revised: 13-May-2024, Manuscript No. JCCLM-24-30863 (R); Published: 20-May-2024 , DOI: 10.35248/2736- 6588.24.7.286
Copyright: © 2024 Ma X. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.