Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2009) Volume 2, Issue 4
In silico analysis of amino acid sequences of some aromatic and aliphatic microbial nitrilases for certain physiochemical properties and specificity to aromatic or aliphatic nitriles has been done. The multiple sequence alignment analysis of amino acid sequences has shown clear differences between aromatic and aliphatic nitrilases in terms of position specific presence of conserved amino acid. Statistical analysis of most of the physiochemical parameters did not show any clear distinction between the two nitrilases. In aromatic nitrilases the conserved amino acid residues besides active site domain triad (Glu, Lys, Cys) were His-129, Asn-168 and Arg-174 and these were replaced by Arg-129, His -168 and Lys-174 in aliphatic nitrilases. The physiochemical properties of these two groups of nitrilases also differed e.g. as compared to aliphatic nitrilases, aromatic nitrilases have lesser number of amino acid residues, lower molecular mass, higher pI values, higher content of Ala and Cys residues.
Keywords: Nitrilase, Aliphatic, Aromatic, Amino acid, Substrate specificity, Multiple alignments.
Nitrilase was the first nitrile metabolizing enzyme reported almost four decades back that converts indole -3- acetonitrile to indole-3-acetic acid (an auxin) in plants (Thimann and Mahadevan, 1964). Later a number of microbes possessing nitrilase activity were isolated which have the capability to metabolize several natural and synthetic nitriles. Nitrilases are the enzymes widely expressed in prokaryotes and eukaryotes that hydrolyze non-peptide carbon-nitrogen bond (Pace and Brenner, 2001). These enzymes have become important tools in green chemistry to replace chemical hydrolysis, oxidation, reduction, etc. As the enzymes are operational at mild reaction conditions and cause least environmental pollution.
Nitrilases differ in substrate specifities and thus find applications in the transformation of a range of nitriles to acids (Banerjee et al., 2002). Initial investigations suggested that nitrilases were specific for aromatic nitriles while nitrile hydratases had affinity for aliphatic nitriles but this distinction has to be reconsidered in the light of rapidly growing information on nitrile degrading enzymes. Now on the basis of substrate specificity, nitrilases can be differentiated as aromatic and aliphatic nitrilases (O’Reilly and Turner, 2003; Banerjee et al., 2002; Mylerova and Martinkova, 2003). It is well known that the sequence of amino acid in a protein determines its structure, function and physiochemical properties (Rabilloud, 2006). How the amino acid sequence of nitrilase/s is related to specificity (aromaticity/aliphaticity) has not been much explored (Yeom et al., 2008).
A number of physiochemical properties e.g. number of amino acid residues, molecular mass, theoretical pI, amino acid composition, negatively charged residues (Asp+Glu), positively charged residues (Arg+Lys), atomic composition, total number of atoms, extinction coefficients (M-1 cm-1) at 280 nm, instability index/ aliphatic index, grand average hydropathicity (GRAVY), etc. of enzymes immensely influence their applications and need to be carefully studied. These properties can be either determined experimentally or deduced from the in silico analysis of amino acid sequences of enzymes available in the databases. Latter approach seems to be attractive for comparison of large number of proteins/enzymes provided the amino acid sequences are available.
In the present communication, we report some physiochemical properties of aliphatic and aromatic nitrilases deduced from the in silico analysis of their amino acid sequences and also the analysis of multiple sequence alignment of some microbial nitrilases which shows that specific type of amino acid at specific positions near the catalytic site are crucial to confer aromaticity or aliphaticity to a nitrilase.
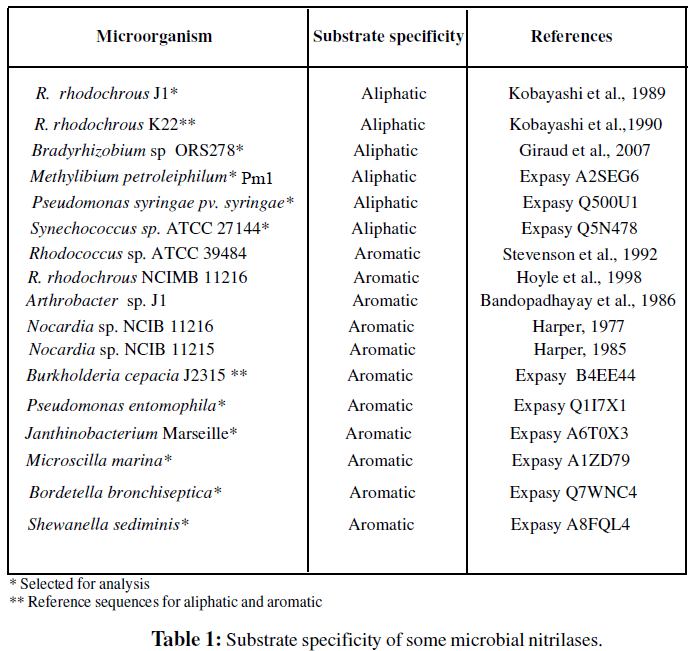
Information about the affinity (aromaticity/aliphaticity) of nitrilases of some microorganisms was obtained from the experimental data earlier published by several researchers (Table 1). Further we had selected amino acid sequences of nitrilase from 12 microorganisms having experimentally proved substrate specificity (Kobayashi et al., 1989; Bhalla et al., 1992; Kobayashi et al., 1989) as well as complete nucleotide sequences and these sequences are not fragmented, pseudo, putative or hypothetical.
The amino acid sequences of the nitrilases were downloaded from the EXPASY proteomic server. Physiochemical data were generated from the Swiss Prot and Expert Protein Analysis System (EXPASY) i.e. the proteomic server of Swiss Institute of Bioinformatics (SIB). FASTA format of sequences were used for analysis.
Blastp (Protein BLAST) was performed to study the homology among the various nitrilase sequences and 6 sequences each belonging to aromatic (E.C.3.5.5.1) and aliphatic (E.C.3.5.5.7) microbial nitrilases were selected (Table 1). All the selected microorganism have complete nucleotide sequences for its nitrilase gene as well as experimentally proved substrate specificity. Clustal W was used for multiple sequence alignment.
Various tools in the Proteomic server {Clustal W, ProtParam, Protein calculator, Compute pI/Mw, ProtScale (Kyte and Dollitle, 1982)} were applied to calculate/deduce different physiochemical properties of nitrilases from the protein sequences.
The molecular weights (Kda) of the various nitrilases were calculated by the addition of average isotopic masses of amino acid in the protein and deducting the average isotopic mass of one water molecule. The pI of nitrilases was calculated using pK values of amino acid according to Bjellqvist et al., (1993).
The atomic composition of nitrilases was derived using the ProtParam tool, available at ExPASy. The extinction coefficient of various nitrilase proteins were calculated using the following equation (Gill et al., 1989): E(Prot) = Numb(Tyr)*Ext(Tyr) + Numb(Trp)*Ext(Trp) + Numb(Cystine)*Ext(Cystine)
The values of aliphatic index of various nitrilase sequences were obtained using ProtParam (ExPASy) tool (Kyte and Doolittle,1982). The instability index and grand average of hydropathicity (GRAVY) were estimated following the method of Guruprasad et al., (1990) and Kyte and Doolittle, (1982), respectively.
Statistical Analysis
An analysis of variance (ANOVA) was conducted on various physiochemical parameter variables for each study with the statistical packages ‘Asistat version-7.4 beta 2008’. F-tests were used to determine the statistical significance. When significant effects were detected, a Tukey test was applied for all pairwise comparisons of mean responses.
Sequence alignment was performed between aliphatic (EC 3.5.5.7) and aromatic nitrilases (EC 3.5.5.1) using Clustal W program (Fig. 1).
Figure 1: Alignment and catalytic triad (Cys 165, lys131,Glu 48) of the amino acid sequences of aliphatic and aromatic nitrilases.(1) Bradyrhizobium sp. ORS278 (2) Methylibium petroleiphilum PM1 (3) Pseudomonas syringae pv. Syringae (4) Rhodococcus rhodochrous K22 (5) Rhodococcus rhodochrous J1 (6) Synechococcus sp. ATCC 27144 / PCC 6301 / SAUG 1402/1 (7) Burkholderia cepacia J2315 / LMG 16656 (8) Pseudomonas entomophila L48 (9) Bordetella bronchiseptica (10) Janthinobacterium Marseille (11) Microscilla marina ATCC 23134 (12) Shewanella sediminis HAW-EB3
In the present studies comparison of amino acid sequences of six nitrilases each under aromatic and aliphatic nitrilase groups to their respective reference sequence (i.e. amino acid sequence of nitrilase of R.rhodochrous K22 for aromatic group of nitrilases and amino acid sequence of nitrilase of Burkholderia cepacia J2315 / LMG 16656 for aromatic group of nitrilases revealed that the selected amino acid sequences were 100% homologous which means these belonged to the same group of nitrilases and were more than 30 % in identity to the reference sequence .The significant
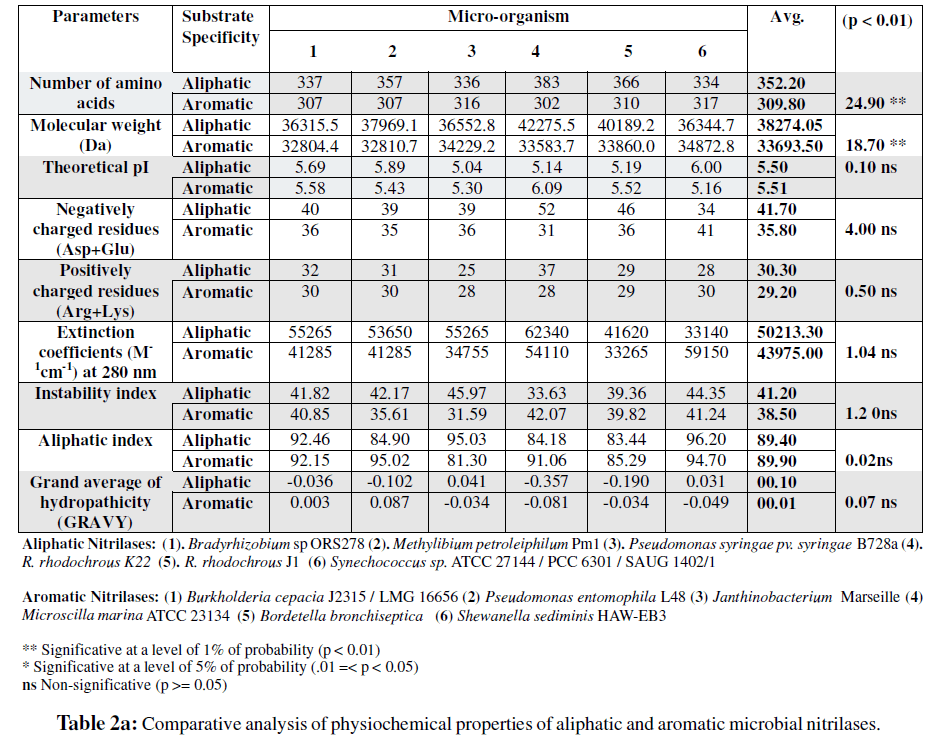
differences in the physiochemical properties of the two types of nitrilases (i.e., aromatic and aliphatic) have been recorded in the present study (Table 2a and Table 2b). The total number of amino acid residues in these nitrilases differed significantly. Aromatic nitrilases have lesser number of amino acid residues (307-317 amino acid) as compared to aliphatic nitrilases (amino acid residues ranging between 334-383). The molecular weight of aromatic nitrilases range between 32804.7-34872.8 and that of aliphatic nitrilases ranges between 36344.7- 42275.5. Theoretical pI varied between 5.16- 6.09 in case of aromatic nitrilases and it was found to be 5.04 - 6.0 for aliphatic nitrilases. It was further observed that the average pI value of aliphatic nitrilases were insignificantly lower than that of aromatic ones studied in the present investigation (Table 2a).
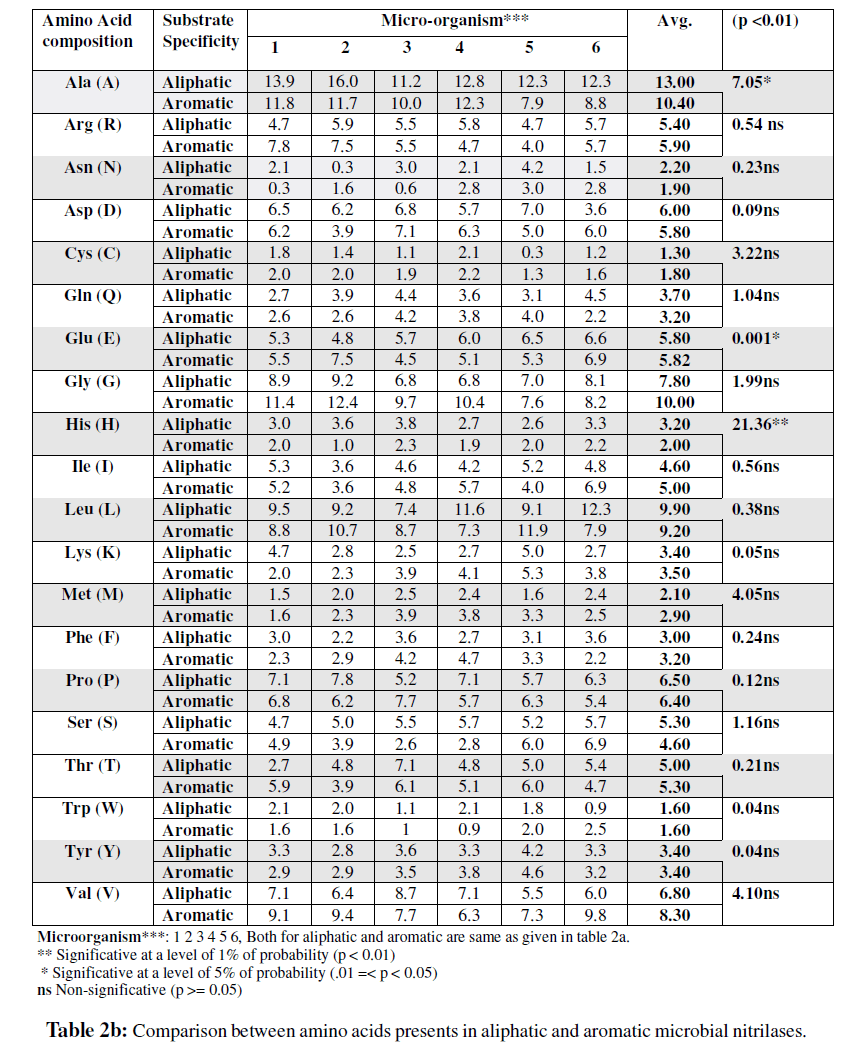
The results of amino acid analysis of nitrilases are presented in Table 2a and 2b. These enzymes contained all the common amino acid. The comparison of the amino acid composition of the aromatic and aliphatic nitrilases has revealed that alanine (Ala), one of the simplest amino acid was found to be the predominant residue in both aromatic and aliphatic nitrilases, however, its percentage was more in aliphatic nitrilases i.e. 1.3 fold higher than aromatic nitrilases. The amino acid cysteine (Cys) is considered to be an important parameter in the calculation of extinction co-efficient of proteins (Gill and von Hippel, 1989) and its content was 1.4 fold higher in aromatic nitrilases. Ala was found to be significantly higher in aliphatic nitrilase. The amino acid Phe, Arg, Gly, Met, Ile, Val and Thr were (1.1, 1.1, 1.3, 1.4, 1.1, 1.2 and 1.1) non significantly higher in aromatic nitrilases while the contents of Gln, Asn, His, Leu, and Ser were (1.2, 1.6, 1.6, 1.1 and 1.2) more in aliphatic nitrilases. No significant difference in percent composition was recorded in Lys, Asp, Pro, Tyr, and Trp (Table 2b). Above results showed that except alanine no other amino acid vary significantly in two groups of nitrilases analyzed in the present studies.
The multiple sequence alignment of aliphatic and aromatic nitrilases exhibited some unique differences between aromatic and aliphatic nitrilases for position specific presence of some amino acid. The position specific (conserved) amino acid in these nitrilases comprised active site domain, and near the N-terminal regions. However, no differences were observed in the conserved amino acid in the C-terminal regions of the enzymes compared in the present study. All nitrilases displayed a conserved catalytic triad consisting of Glu-48, Cys-165 and Lys-131. The other conserved amino acid residues in both aliphatic and aromatic nitrilases included Ala -11, 199, 215 and 278; Gln- 14; Pro-47, 259 and 55; Glu-48, 106, 138, 167 and 217; Tyr- 54, 106 and 298; Arg-130, 139 and 300; Lys- 131 and 261; Gly- 103, 145 and 296; Cys-165; Trp-166; Val-222; Thr-135; Asp- 281, 293 and 302; His- 297 (Fig.1). The conserved amino acid residues present in aromatic nitrilases comprised His- 129, 202 Tyr-142, 196 Asp-146, Met-170, 200 Arg-174, 208, Pro-188, Thr-189, Asp-191, Gly-66, 252 while Pro-17, Ala-36, 37, 92, Arg- 94, 129 , Ile-119, His- 168, 185 Glu-182, 273, Asp- 239, Gly-250 emerged as conserved amino acid in aliphatic nitrilases (Fig.1).
Negative charge residues (Asp and Glu) were found to be higher in aliphatic nitrilases i.e 41.66 in aliphatic nitrilases and 35.83 in aromatic nitrilases. Insignificant difference was observed between aromatic and aliphatic nitrilases for positively charged amino acid residues. Instability indices of these two groups of nitrilases indicated that aromatic nitrilases were more stable as compared to aliphatic nitrilases (Table 2a). The aliphatic index values and values for grand average hydropathicity (GRAVY) were higher for aliphatic nitrilases than aromatic nitrilases compared in the present study.
In the present study an attempt has been made to differentiate two groups of nitrilases i.e. aromatic nitrilases and aliphatic nitrilases on the bases of their amino acid sequences and physiochemical properties. The selected sequences in each group were 100 % homologous and more than 30 % identical with respective to model sequence of each group i.e. aliphatic nitrilase amino acid sequence of Rhodococcusrhodochorus K22 (Q02068,UniProtKB/Swiss-Prot) and aromatic nitrilase amino acid sequence of Burkholderia cepacia J2315 / LMG 16656 (B4EE44,UniProtKB/Swiss- Prot). Significant differences in total number of amino acid residues between aliphatic nitrilases and aromatic nitrilases indicated that total number of amino acid and molecular weight might be playing some role in providing the substrate specificity to these two groups of nitrilases.
The two groups of nitrilases did not show significant differences for pI values. This property of the protein seems not to be related with the substrate specificity of the nitrilase enzymes considered in the present study. The present study has further revealed that specificity of aliphatic nitrilases towards aliphatic nitriles might be due to higher number of negatively charged amino acid residues and higher aliphatic index values than that of aromatic nitrilases.
Due to diversity of 20 amino acid, and to the incredible number of combinations they afford, proteins differ widely in physiochemical properties as well as in substrate specificity (O’Reilly and Turner, 2003; Brenner, 2003; Yeom et al., 2008). The results of this study has also confirmed that amino acid number and their percent composition in nitrilases significantly affect the substrate specificity i.e. aromaticity and aliphaticity of these enzymes. According to Kobayashi et al., (1989) Ala is the predominant amino acid in the nitrilases and this has also been confirmed in amino acid
sequences of compared nitrilases in the present investigation. The observation that Ala content is comparatively higher in aliphatic nitrilases led us to infer that higher content of Ala might be contributing towards specificity of nitrilases towards aliphatic nitriles. Another important amino acid
which was found to play a crucial role in nitrilases is Cys (Kobayashi et al., 1992) which generally imparts stability to proteins due to the formation of disulphide bonds. The result in present study also indicated that aromatic nitrilases are more stable as is shown by the instability index values (Table 2a) of various nitrilases considered in present study. This observation is further supported by the fact that aromatic nitrilases have higher percentage of Cys content than the aliphatic nitrilases. Cys is also the part of reaction centre in nitrilases and attack the cyno group of nitrile as a nucleophile (Yeom et al., 2008; Kobayashi et al., 1992).
The present investigation has led to find out very clear differences between aromatic and aliphatic nitrilases in terms of position specific presence of certain amino acid. Amino acid residues at position 129, 168 and 174 in aromatic nitrilases are invariable His, Asn and Arg respectively while these positions are occupied by Arg, His and Lys respectively in aliphatic nitrilases. In the aromatic nitrilases, Met- 170 is conserved while Glu-182 and His-185 are conserved in aliphatic nitrilases. The position specific amino acid residues might be playing important role in conferring differential substrate specificity to aromatic and aliphatic nitrilases. Importance of site specific amino acid residues in relation to substrate specificity of nitrilases of Rhodococcus rhodochrous ATCC 33278 has been highlighted in a recent publication (Yeom et al., 2008). According to these authors Tyr-142 in nitrilases of Rhodococcus rhodochrous ATCC 33278 is responsible for its affinity towards aromatic nitrilases.
The relationship predicted with substrate specificity and position specific amino acid residues of aromatic and aliphatic nitrilases can only be validated by in vitro experiments involving site directed mutagenesis to substitute the conserved amino acid residue/s of aromatic nitrilases with the conserved amino acid reisdue/s of aliphatic nitrilase orvice-versa to see whether one or any or many of the predicted amino acid substitution leads to change substrate specificity of nitrilases from aromatic nitriles to aliphatic nitriles or vice-versa.
A number of physiochemical properties of aliphatic and aromatic microbial nitrilases have been deduced from amino acid sequences of some microbial nitrilases. The aromatic and aliphatic nitrilases mainly differ in the total number of amino acid, molecular weights and composition of amino acid. This study has clearly shown differences between these two types of nitrilases in conserved amino acid residues at several positions. The results of the present investigation will be quite useful in prediction and selection of aromatic/ aliphatic nitrilases from hither to reported nitrilases or from the large number of sequenced microbial genomes.
Authors are grateful to the Department of Biotechnology Govt. of India, New Delhi, India for financial support to the Bioinformatics Centre. Rekha Kushwaha is thankful to UGC for financial support in the form of Dr. D.S. Kothari Post Doctoral Fellowship.