Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2023)Volume 16, Issue 3
Single nucleotide polymorphism play a vital role in understanding the genetic basis of numerous complex human diseases but the identification of functional SNPs in a disease related gene still continuous to be a major challenge. In spite of the fact that recent advances in DNA sequencing techniques have led to an increase in the identification of SNPs in human BRCA genes, further information regarding the deleterious probability of many variants is not available for those SNPs classified as ‘variants of unknown significance’ and ‘variants with conflicting interpretations of pathogenicity’. The objective of this study was to analyze non synonymous coding SNPs of human BRCA2 gene stated as ‘SNPs with conflicting interpretations of pathogenicity’ in NCBI by in silico method in order to predict deleterious SNPS which may potentially be manipulated for diagnostic and management purposes of different pathologic conditions. Human BRCA2 gene SNPs were retrieved from the National Center for Biotechnology Information (NCBI) database, dbSNP GeneMANIA ver. 3.1.2.8 was applied to study interaction of BRCA2 with other genes. Sorting Intolerant From Tolerant (SIFT) online software was applied for the in silico prediction of functional effects of the BRCA2 coding missense SNPs and the molecular phenotypic effects of selected deleterious mutant SNPs on the resultant protein were analyzed by using hope online server. 302 non synonymous coding SNPs were retrieved from NCBI out of which 88 were found to deleterious with SIFT score of <0.05. 20 of the deleterious SNPs were very deleterious with SIFT score of 0.0. The phenotypic effect of selected very deleterious SNPs was predicted by project hope web server and the findings suggest that these SNPs can be correlated with different human clinical conditions.
In silico analysis; BRCA2; Single nucleotide polymorphisms; National Center for Biotechnology Information (NCBI); Sorting Intolerant from Tolerant (SIFT)
Single Nucleotide Polymorphisms (SNPs) are known to play a vital role in understanding the genetic basis of numerous complex human diseases [1]. For instance, the genetic bases of human phenotypic variation could be understood by knowing the functions of various SNPs. But the identification of functional SNPs in a disease related gene still continuous to be a major challenge. In spite of the fact that recent advances in DNA sequencing techniques have led to an increase in the identification of SNPs in human BRCA genes, further information regarding the deleterious probability of many variants is not available [2].
Hereditary breast and ovarian cancer cases are partly known to be attributed to heritable mutations in the BRCA (BRCA1 and BRCA2) tumor suppressor genes which tremendously increase the chance of cancer development in individuals who carry these mutations. Characteristically, BRCA2 gene has 27 exons collectively encoding the BRCA2 protein which is composed of 3,418 amino acids. The N terminal domain of BRCA2 harbors a transcription activating domain (residues 18–105) and the middle region is composed of eight conserved repeated motifs which are essential for binding with RAD51 gene for consequent initiation of DNA repair [3]. A conserved DNA binding domain is also located in the C terminal region which interacts with several proteins including DSS1.
The involvement of BRCA2 gene in homologous recombination and its paramount role in cancer susceptibility is well documented in literature with heterozygous pathogenic germ line mutations of the gene being responsible for Hereditary Breast and Ovarian Cancer syndrome (HBOC) [4]. BRCA2 mutation is also implicated to increase susceptibility to other cancer types such as prostate or pancreatic cancers; BRCA2 germ line mutations also cause fanconi anemia group D1 syndrome which is commonly accompanied by an increased risk of various oncological conditions such as leukemia, glioma, etc.
BRCA2 gene generally plays a vital role in maintaining the stability of genetic information in cells; its mutation and consequent abnormal expression thus leads to the destruction of normal structure of cells which leads to the growth of tumor cells.
The integration of large data sets and algorithms is crucial in our understanding of cancer. The use of in silico models is helpful to alleviate difficulties posed by data complexity. Besides, in silico analyses can also help to disclose hidden underlying molecular phenomena [5].
Advances in bioinformatics and computational biology have brought about the development of many in silico tools which have been employed by scientists in different areas of science including biology and genetics in simulative genetic, molecular studies such as assessment of mutational variants of different genes. The incorporation of tremendous mathematical algorithms, physical theories, biochemical concepts in to computer systems has made available the possibility of generating meaningful scientific ideas using the computer systems such computations may in fact produce information not available in actual experiments. Several branches of computer based works have, therefore, been developed one of which is computational biology. Several works involving such computations are being reported in literature in the medical field an example of which is computer aided drug design. New emergent journals are also being established which are engaged in publishing special in silico based studies following such developments. The usage of in silico studies has strong impact on the identification of candidate SNPs since such studies are easy and less costly and can facilitate future genetic studies.
Numerous pathogenic mutations and benign variants are already reported covering different regions of the human BRCA2 gene in the ClinVar database. Such mutations which shorten the encoded BRCA2 protein through various mutational mechanisms including: Frame shift, nonsense codon insertion, or altered splicing are mainly settled to be considered as pathogenic. Nevertheless, more than 50% of BRCA2 mutations described in ClinVar are known to be missense mutations causing non synonymous alterations in the BRCA2 protein, nearly 90% of which being categorized as either Variants of Unknown Significance (VUS) or variants with conflicting interpretations of pathogenicity.
These types of variants pose a major clinical challenge for cancer risk management in mutation carriers. It is not uncommon, though, to encounter such missense Single Nucleotide Polymorphisms (SNPs) which are linked to different pathologies at evolutionarily conserved regions which have a detrimental role both at the structural and functional levels of the protein.
Therefore, this study focused on the analysis of missense codding SNPs of BRCA2 with conflicting interpretations of pathogenicity [6]. Previous studies including have conducted a computational prediction of possible deleterious functional SNPs in human BRCA2 gene but clearly additional online submissions of human BRCA2 SNPs have been made after this study, hence it is reasonable to do an Insilco study with added data and newer tools and approaches. The aim of this study is to analyze non synonymous coding SNPs of human BRCA2 by in silico method and to determine the effect of these SNPs on the molecular and phenotypic features of the mutated BRCA2 protein which may be linked with some pathology [7].
Objective
The objective of this study was to analyze non synonymous coding SNPs of human BRCA2 gene stated as ‘SNPs with conflicting interpretations of pathogenicity’ in NCBI by in silico method in order to predict deleterious SNPS which may potentially be manipulated for diagnostic and management purposes of different pathologic conditions.
Hypothesis
Studies addressing BRCA2 mutations from a bioinformatics point of view have been published and have identified significant SNPs. But continued in silico analysis of BRCA2 SNPs by incorporating recently submitted data and newer bioinformatics tools may be important to detect novel significant SNPs [8]. The acquired knowledge may be used as a starting point for the development of new, useful SNP markers for medical testing and therapeutic practices aimed at treating different human malignancies which involve this gene.
Data mining
Human BRCA2 gene SNPs were retrieved from the National Center for Biotechnology Information (NCBI) database, dbSNP. BRCA2 was used as search term and filters were used to narrow down search results into two categories: Missense SNPs with conflicting interpretations of pathogenicity.
Outlining gene–gene interactions
GeneMANIA version. 3.1.2.8 was applied to study interaction of BRCA2 with other genes. Results from such studies can be utilized in haplotype studies. This server finds other genes that are related to a set of input genes, using a very large set of functional association. GeneMANIA is a flexible, user friendly web interface which can be used to generate presumptions or hypotheses about the function of genes; it analyzes gene lists and prioritizes them for functional assays. GeneMANIA also reports weights that indicate the predictive value of each selected data set for the query. Six organisms are currently supported in this interface (Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Homo sapiens and Saccharomyces cerevisiae) [9]. Users of this web interface can select arbitrary subsets of the data sets associated with the organism of their choice to perform their analyses and can upload their own data sets to analyze. The high accuracy of the GeneMANIA prediction algorithm, an intuitive user interface and large database make this bioinformatics tool a useful aid for biologists.
Evaluation of non–synonymous coding single nucleotide polymorphisms of BRCA2
In silico prediction of functional effects of the BRCA2 coding missense SNPs was carried out using Sorting Intolerant From Tolerant (SIFT) online software to classify them into neutral/deleterious and a probability is calculated based on that prediction a tolerance index of <0.05 is considered deleterious. This software presumes that important amino acid changes at well conserved positions in the protein family are liable to be predicted as deleterious [10]. To predict whether an amino acid substitution in a protein will affect protein function, SIFT considers the position at which the change occurred and the type of amino acid change.
Given a protein sequence, SIFT chooses related proteins and obtains an alignment of these proteins with the query sequence. Based on the amino acids appearing at each position in the alignment, SIFT calculates the probability that an amino acid at a position is tolerated conditional on the most frequent amino acid being tolerated [11]. If this normalized value is less than a cutoff (0.05), the substitution is predicted to be deleterious.
Predicting the molecular phenotypic effects of deleterious single nucleotide polymorphism
The molecular phenotypic effects of selected deleterious mutant SNPs on the resultant protein were analyzed by using hope online server. Mutants were selected for analysis via HOPE server based on protein residue position. Hope online mutant analysis server is a fully automatic program that analyzes the structural and functional effects of point mutations. It does the analysis by collecting information from various sources including calculations on the 3D coordinates of the protein by using what if Web services, sequence annotations from the UniProt database, and predictions by DAS services. Homology models are built with YASARA [12]. Data is stored in a database and used in a decision scheme to identify the effects of a mutation on the protein’s 3D structure and function. Hope builds a report with text, figures, and animations that is easy to use and understandable for researchers. For proteins with no structural information and solved 3D structure, hope uses annotations from the UniProt database and predictions from the Reprof software for mutational analysis (Figure 1).
Figure 1: Flow chart of BRCA2 in silico analysis process.
Data mining
302 coding non synonymous BRCA2 SNPs were downloaded from National Center for Biotechnology Information (NCBI) database, dbSNP for analysis by different bioinformatics tools.
Outlining gene–gene interactions via geneMANIA
BRCA2 gene interacts with different genes while performing different cellular functions such as DNA recombination, recombinational repair, double strand break repair and many more which are majorly targeting in tackling tumor development [13]. Figure 2 shows the gene–gene interactions of human BRCA2 gene.
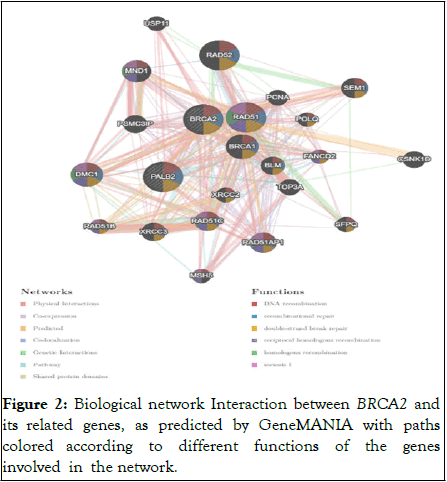
Figure 2: Biological network Interaction between BRCA2 and its related genes, as predicted by GeneMANIA with paths colored according to different functions of the genes involved in the network.
Predicting effects of non sinonymous single nucleotide polymorphisms by sift online software
Out of 302 non synonymous coding BRCA2 SNPs retrieved from NCBI (book 1.zip), 88 SNPs (book 2.zip) were predicted to be deleterious according to SIFT online software with SIFT scores <0.05. Out of the deleterious SNPs, 23 SNPs had a score of 0.00 (Table 1).
| SNP | CHR 13 | AA change | Gene | Region | Score | Prediction |
|---|---|---|---|---|---|---|
| rs80358726 | 13 | Pl68T | BRCA2 | CDS | 0 | Deleterious |
| rs80358925 | 13 | P2329L | BRCA2 | CDS | 0 | Deleterious |
| rs80358974 | 13 | I2495T | BRCA2 | CDS | 0 | Deleterious |
| rs80358995 | 13 | F2562L | BRCA2 | CDS | 0 | Deleterious |
| rs80359009 | 13 | G2609A | BRCA2 | CDS | 0 | Deleterious |
| rs80359010 | 13 | D26 1IG | BRCA2 | CDS | 0 | Deleterious |
| rs80359029 | 13 | Y2660D | BRCA2 | CDS | 0 | Deleterious |
| rs80359076 | 13 | R2784Q | BRCA2 | CDS | 0 | Deleterious |
| rs80359136 | 13 | Q2925H | BRCA2 | CDS | 0 | Deleterious |
| rs80359150 | 13 | R299 1H | BRCA2 | CDS | 0 | Deleterious |
| rs80359171 | 13 | R3052Q | BRCA2 | CDS | 0 | Deleterious |
| rs80359246 | 13 | P3280H | BRCA2 | CDS | 0 | Deleterious |
| rs200210279 | 13 | S3291C | BRCA2 | CDS | 0 | Deleterious |
| rsll571769 | 13 | A2951T | BRCA2 | CDS | 0 | Deleterious |
| rs80358726 | 13 | Pl68A | BRCA2 | CDS | 0 | Deleterious |
| rs80358730 | 13 | Kl69R | BRCA2 | CDS | 0 | Deleterious |
| rs80358921 | 13 | R23 18Q | BRCA2 | CDS | 0 | Deleterious |
| rs80358978 | 13 | G2508S | BRCA2 | CDS | 0 | Deleterious |
| rs80359014 | 13 | 12627F | BRCA2 | CDS | 0 | Deleterious |
| rs80359105 | 13 | R2842L | BRCA2 | CDS | 0 | Deleterious |
| rs80359129 | 13 | G2901D | BRCA2 | CDS | 0 | Deleterious |
| rs80359150 | 13 | R299 1H | BRCA2 | CDS | 0 | Deleterious |
| rs13507014 | 13 | D3341E | BRCA2 | CDS | 0 | Deleterious |
Table1: BRCA2 SNPS with SIFT score of 0.00.
Predicting the effect of selected deleterious SNPs on the phenotypic properties of BRCA2 protein
The effect of 7 selected very deleterious SNPs with sift score of 0.00 on the structure, conservation and amino acid properties of the mutated BRCA2 protein was predicted via hope online software by analyzing the amino acid changes at specified regions of BRCA2 protein according to the predictions by the SIFT software. Hope predictions for 7 selected most deleterious SNPS (SIFT score=0.00) are summarized below [14].
rs80358974
The mutation of isoleucine into a threonine amino acid at position 2495 due to this SNP is shown in the following Figure 3.
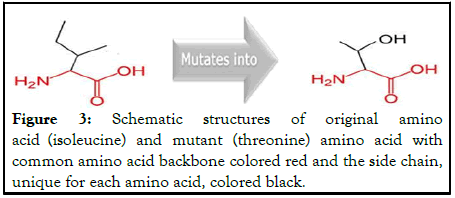
Figure 3: Schematic structures of original amino acid (isoleucine) and mutant (threonine) amino acid with common amino acid backbone colored red and the side chain, unique for each amino acid, colored black.
The characteristics of this mutation and the hope predicted molecular and phenotypic effects are summarized in the following Table 2.
| No. | Characteristics of the mutation | Hope predicted associated molecular and phenotypic features |
|---|---|---|
| 1 | Mutant residue smaller than wild type residue. | Disturbance of domain’s the core structure. |
| 2 | Mutation located in a specially annotated region in UniProt: Interaction with FANCD2. | Regional functional disturbance leading to possible genomic instability. |
| 3 | Mutation occurs in a 100% conserved residue. | Mutation is damaging to the protein. |
| 4 | Mutant residue les hydrophobic than wild type residue. | Mutation cause an empty space in the core of the protein with loss of hydrophobic interactions. |
Table 2: Summary of mutation characteristics and phenotypic effects of SNP rs80358974.
The three dimensional views of the wild type and mutant BRCA2 proteins are shown in the following Figure 4.
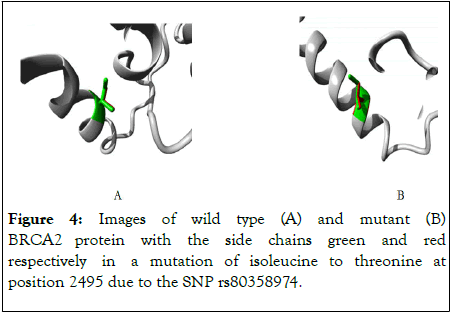
Figure 4: Images of wild type (A) and mutant (B) BRCA2 protein with the side chains green and red respectively in a mutation of isoleucine to threonine at position 2495 due to the SNP rs80358974.
rs80358995
This SNP results in the mutation of a phenylalanine into a leucine at position 2562 (Figure 5).
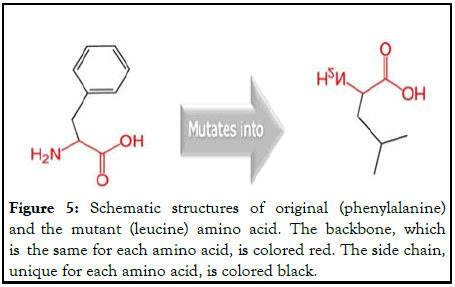
Figure 5: Schematic structures of original (phenylalanine) and the mutant (leucine) amino acid. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the hope predicted molecular and phenotypic effects are summarized in the following Table 3.
| No. | Characteristics of the mutation | Hope predicted molecular and phenotypic features |
|---|---|---|
| 1 | Mutant residue smaller than wild type residue. | An empty space created in the core of the protein, disturbance of the domain’s core structure. |
| 2 | Mutation located in a specially annotated region in UniProt: Interaction with SEM1. | Disturbance of regional function which may lead to impaired cellular functions associated with inability to remove misfolded/damaged proteins. |
| 3 | Mutation occurs in a 100% conserved residue. | Mutation is damaging to the protein. |
| 4 | Mutant residue more hydrophobic than wild type residue. | Disturbed hydrophobic interaction of mutant residue; hydrogen bond formation is also affected. |
Table 3: Summary of mutation characteristics and phenotypic effects of SNP rs80358995.
The following Figure 6 shows close up of 3D view of the mutation.
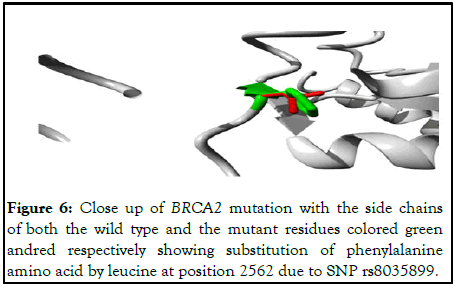
Figure 6: Close up of BRCA2 mutation with the side chains of both the wild type and the mutant residues colored green andred respectively showing substitution of phenylalanine amino acid by leucine at position 2562 due to SNP rs8035899.
rs80359009
This SNP induces a mutation of a glycine into an alanine at position 2609 (Figure 7).
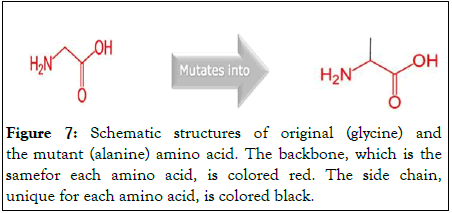
Figure 7: Schematic structures of original (glycine) and the mutant (alanine) amino acid. The backbone, which is the samefor each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the hope predicted molecular and phenotypic effects are summarized in the following Table 4.
| No. | Mutation characteristic | Hope predicted molecular and phenotypic features |
|---|---|---|
| 1 | Mutation of a glycine into another amino acid (alanine). | Loss of protein function associated with inflexibility of incorporated non glycine amino acid. |
| 2 | Mutant residue bigger than wild type residue. | Inability of mutant residue to fit into a structure leading to disturbance of core structure of domain. |
| 3 | Mutation located in a specially annotated region in UniProt: Interaction with SEM1. | Disturbance of regional function which may lead to impaired cellular functions associated with inability to remove misfolded/damaged proteins. |
| 4 | Mutation occurs in a 100% conserved residue. | Mutation is damaging to the protein. |
| 5 | Mutant residue more hydrophobic than wild type residue. | Disturbed hydrophobic interaction of mutant residue. |
Table 4: Summary of mutation characteristics and phenotypic effects of SNP rs80359009.
The 3D view of the wild type and mutant BRCA2 proteins is depicted in the following Figure 8.
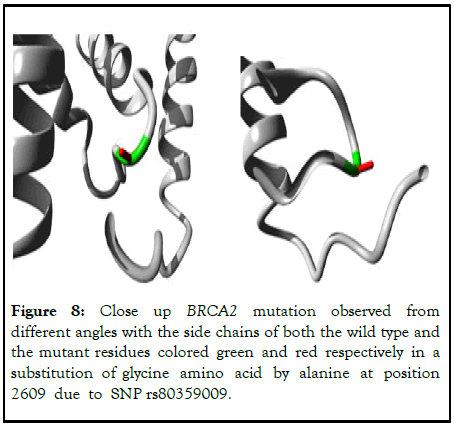
Figure 8: Close up BRCA2 mutation observed from different angles with the side chains of both the wild type and the mutant residues colored green and red respectively in a substitution of glycine amino acid by alanine at position 2609 due to SNP rs80359009.
rs80359010
The mutation of aspartic acid into glycine at position 2611 due to this SNP is schematically represented in Figure 9.
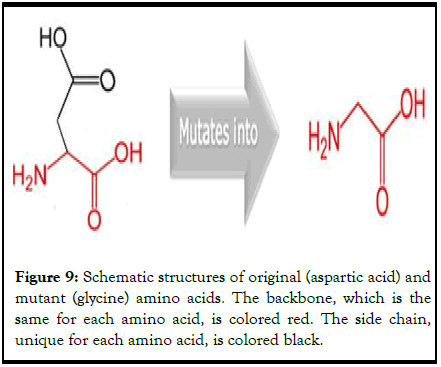
Figure 9: Schematic structures of original (aspartic acid) and mutant (glycine) amino acids. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the HOPE predicted molecular and phenotypic effects are summarized in the following Table 5.
| No. | Mutation characteristic | Hope predicted molecular and phenotypic features |
|---|---|---|
| 1 | Mutation of an aspartic acid in to a glycine amino acid. | Positional disturbance of protein rigidity due to introduction of a flexible glycine amino acid. |
| 2 | Mutant residue smaller than wild type residue. | Mutant residue is not in the correct position to make the same hydrogen bond as the original wild type residue leading to structural disturbance. |
| 3 | Mutation located in a specially UniProt annotated region: Interaction with SEM1. | Disturbance of regional function which may lead to impaired cellular functions associated with inability to remove misfolded/damaged proteins. |
| 4 | Mutation occurs in a 100% conserved residue. | Mutation is damaging to the protein. |
| 5 | Mutant residue more hydrophobic than wild type residue. | Disturbed hydrophobic interaction of mutant residue; hydrogen bond formation is also affected. |
| 6 | Wild type’s charge is negative while mutant is neutral. | Disturbed ionic interaction which may lead to loss of interactions with other molecules |
Table 5: Summary of mutation characteristics and phenotypic effects of SNP rs80359010.
Image of ribbon presentation of the mutated BRCA2 protein is displayed in the following Figure 10.

Figure 10: Overview BRCA2 protein in ribbon presentation with the side chain of the mutated residue colored magenta and shown as small balls during a substitution of aspartic acid by glycine amino acid at position 2611 due to SNP rs80359010.
This is a mutation of a tyrosine into an aspartic acid at position 2660 (Figure 11).
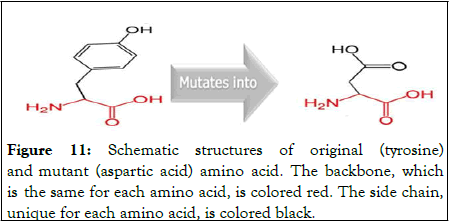
Figure 11: Schematic structures of original (tyrosine) and mutant (aspartic acid) amino acid. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the hope predicted molecular and phenotypic effects associated with this SNP are summarized in the Table 6.
| No. | Mutation characteristic | Hope predicted molecular and phenotypic effects |
|---|---|---|
| 1 | Mutant residue is smaller than wild type residue. | Mutant residue is not in the correct position to make the same hydrogen bond as the original wild type residue did. |
| 2 | Mutation located in a specially annotated region in UniProt: Interaction with SEM1. | Disturbance of regional function which may lead to impaired cellular functions associated with inability to remove misfolded/damaged proteins. |
| 3 | Mutation occurs in a 100% conserved residue | The mutation is damaging to the protein. |
| 4 | Mutant residue more hydrophobic than wild type residue | Difference in hydrophobicity affects hydrogen bond formation; mutant residue may not be able to form a hydrogen bond with aspartic acid at position 2611. |
| 5 | Wild type’s charge is neutral while mutant is negatively charged | The mutant residue introduces a charge in a buried residue which can lead to protein folding problems. |
Table 6: Summary of mutation characteristics and phenotypic effects of SNP rs80359029.
3D view of the mutational effect on the protein is depicted in the following Figure 12.
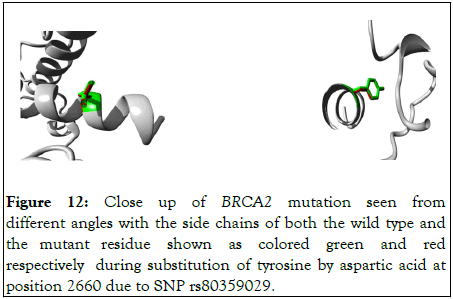
Figure 12: Close up of BRCA2 mutation seen from different angles with the side chains of both the wild type and the mutant residue shown as colored green and red respectively during substitution of tyrosine by aspartic acid at position 2660 due to SNP rs80359029.
rs80358730
A mutation of glycine amino acid into a serine amino acid at position 2508 is depicted in the following Figure 13.
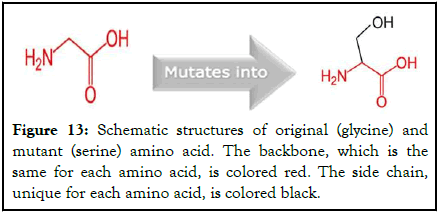
Figure 13: Schematic structures of original (glycine) and mutant (serine) amino acid. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the hope predicted molecular and phenotypic effects associated with this SNP are summarized in the following Table 7.
| No. | Mutation characteristic | Hope predicted molecular and phenotypic features of mutants |
|---|---|---|
| 1 | Mutation of a glycine into another amino acid (serine). | Loss of protein function and structural disturbance associated with inflexibility of incorporated non glycine amino acid. |
| 2 | Mutant residue is bigger than wild type residue. | Inability of mutant residue to fit into a structure leading to disturbance of core structure of domain. |
| 3 | Mutation located in a specially UniProt annotated region: Interaction with FANCD2. | Regional functional disturbance leading to possible genomic instability. |
| 4 | Mutation occurs in a 100% conserved residue. | The mutation is probably damaging to the protein. |
| 5 | Mutant residue more hydrophobic than wild type residue. | Disturbed hydrophobic interaction of mutant residue; hydrogen bond formation is also affected. |
| 6 | Wild type’s charge is neutral while mutant is negatively charged. | The mutant residue introduces a charge in a buried residue which can lead to protein folding problems. |
Table 7: Summary of mutation characteristics and phenotypic effects of SNP rs80358730.
Figure 14 shows close up of the 3D view of the mutation.
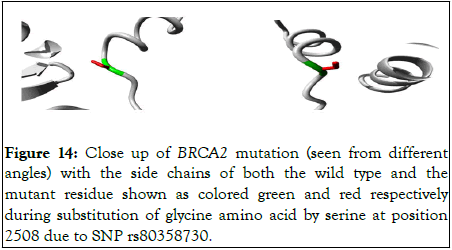
Figure 14: Close up of BRCA2 mutation (seen from different angles) with the side chains of both the wild type and the mutant residue shown as colored green and red respectively during substitution of glycine amino acid by serine at position 2508 due to SNP rs80358730.
rs80359014
The mutation an isoleucine into phenylalanine at position 2627 associated with this specific SNP is shown in Figure 15.
Figure 15: Schematic structures of original (Isoleucine) and the mutant (Phenylalanine) amino acid. The backbone, which is the same for each amino acid, is colored red. The side chain, unique for each amino acid, is colored black.
The characteristics of this mutation and the HOPE predicted molecular and phenotypic effects associated with this SNP are summarized in the following Table 8.
| No. | Mutation characteristic | Hope predicted molecular and phenotypic effects |
|---|---|---|
| 1 | Mutant residue is bigger than the wild type residue. | Inability of mutant residue to fit into a structure leading to disturbance of core structure of domain. |
| 2 | Mutation located in a specially annotated region in uniprot: Interaction with SEM1. | Disturbance of regional function which may lead to impaired cellular functions associated with inability to remove misfolded/damaged proteins. |
| 3 | Mutation occurs in a 100% conserved residue. | The mutation is probably damaging to the protein. |
Table 8: Summary of mutation characteristics and phenotypic effects of SNP rs80359014.
The Figure 16 shows close up of the mutation observed from different angles.
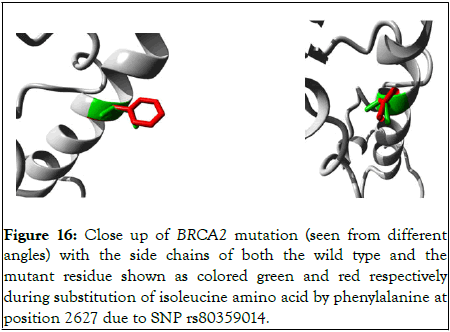
Figure 16: Close up of BRCA2 mutation (seen from different angles) with the side chains of both the wild type and the mutant residue shown as colored green and red respectively during substitution of isoleucine amino acid by phenylalanine at position 2627 due to SNP rs80359014.
Different branches of computer based works have been developed as part of the advancement of science and technology among which is computational biology [15]. Although computational results may not be 100% correlated with experimental results, considerable dedications have been made by researchers in the last few decades to perform computer based works in the prediction and interpretation of experimental achievements. This is due to the fact that different computational analyses can provide researchers with valuable information which may not be experimentally acquired [16]. The value of in silico analysis of Single Nucleotide Polymorphisms (SNPs) for the prediction of variants which are significantly associated with disease has been appreciated recently and this may facilitate future association studies. The approach has been practiced for many disorders, especially being utilized for cancer related genes [17].
The current study focused on the analysis of non-synonymous coding SNPs of BRCA2. The analysis utilized different bioinformatics tools (Figure 1) and involved 302 SNPs from the dbSNP data base in NCBI. 88 (29.2%) of the data base retrieved SNPs were predicted to be deleterious by SIFT with tolerance index score of <0.05; 23 of these were extremely deleterious with tolerance index score of 0.0.
The percentage of BRCA2 deleterious SNPs predicted by SIFT tool in the current study is less than the percentage of deleterious SNPs reported by Poon [18]. Utilizing the same tool which was 51.48%.This difference may be due to the larger sample size employed for analysis by the previous study. There is also a difference in the classification of SNPs in that the previous study analyzed SNPs all of which were classified as ‘Variants of Unknown Significance (VUS)’ in NCBI but our data incorporated SNPs reported in NCBI as ‘variants with conflicting interpretations of pathogenicity’. 23 of the SIFT predicted SNPs in the current study were extremely deleterious having had a tolerance index of 0.0. Out of these molecular and phenotypic effects on the resultant BRCA2 was predicted for 7 selected SNPs by making used of the hope server. 3D images were not possible to be generated for some SNPs through hope and thus they were not chosen. Besides, SNPs occurring at longer positions were not convenient for HOPE analysis. We believe the HOPE analyzed SNPs in this study could indicate the pattern of the phenotypic effect deleterious BRCA2 SNPs have on the mutant BRCA2 protein [19].
The biologically important characteristics of the mutations predicted in these SNPs can be summarized as: Replacement of a glycine amino acid by another amino acid; incorporation of a glycine amino acid in to a residue; size difference between wild type and mutant residues; mutation being located in a special region of the protein where there is interaction with another gene; mutation happening in a 100% conserved residue; hydrophobicity difference between wild type and mutant residues; and a difference of charge between wild type and mutant residues.
Mutation of a glycine into another amino acid, alanine at position 2609 was predicted by HOPE for the SNP rs80359009. This has mainly a structural effect [20]. Glycine being a flexible molecule adds to the flexibility of the wild type protein. The small size of glycine gives it the following cellular functions: A flexible link in proteins allowing for the formation of helices, as an extracellular signaling molecule, a recognition site on cell membranes and enzymes, a modifier of molecular activity via conjugation and extension of hormone precursors, and an osmoprotectant. These cellular attributes will be lost in the mutant protein due to incorporation of a less flexible molecule, alanine. This will further lead to altered cellular signaling and interaction properties [21]. The effect of this phenotypic variation in the development of BRCA related human clinical conditions needs further functional and association studies.
The incorporation of a glycine amino acid in to a residue replacing another amino acid, aspartic acid at BRCA2 position 2611 was predicted in the SNP rs80359010. This mutation removes the less flexible amino acid and replaces it with a more flexible amino acid, glycine. This may lead to positional disturbance of the rigidity of the wild type protein creating a mutant protein with undesirable signaling and interaction properties (due to the removal of glycine) at this position which may interfere with the DNA repair and tumor suppression properties of BRCA2 which it does by interacting with other genes.
A difference of size between wild type and mutant proteins was predicted in all of the HOPE analyzed most deleterious SNPs of BRCA2. The mutant protein was smaller than the wild type in 4 SNPs (rs80358974, rs80358995, rs80359010, and 80359029) while the mutant protein was bigger than the wild type protein in the rest 3 SNPs (rs80359009, rs80358730, and rs80359014) [22]. In both cases the consequential structural effect is disturbance of the protein domain’s core structure. This also may render the mutant BRCA2 protein defective in attaining the interactive ability to involve in DNA repair and tumor suppression. It is known that cancer causing missense mutations of BRCA2 have the ability to disrupt an intracellular protein assembly mechanism thereby disabling genome maintenance. It is also well documented that BRCA2 plays a vital role in upholding the integrity of the human genome via different biological processes including; localization and assembly of RAD51 (a recombination enzyme) on DNA substrates during DNA repair by Homologous DNA Recombination (HDR); protecting the structure of delayed DNA replication forks; suppressing spontaneous RNA-DNA hybrid (R loop) formation through nuclear RNA export regulation or promoter proximal RNA polymerase II pausing; and involving in events leading to chromosome segregation. These and other related cellular processes may be halted due to the aforementioned structural disturbances associated with the deleterious single nucleotide polymorphisms of BRCA2.
All the 7 mutations occurred in a specially annotated region of BRCA2 by uniprot, a region where there is interaction with other genes: 5 mutations (rs80358995, rs80359009, rs80359010, rs80359029, rs80359014) occurred in a region where the BRCA2 gene interacts with SEM1/DSS1 gene and the 2 mutations (rs80358974, rs80358730) occurred in a region where it interacts with FANCD2 gene.
The carboxyl terminal domain of BRCA2 also called DNA Binding Domain (DBD) is known to bind to the small acidic residue SEM1 also called DSS1 with BRCA2 DSS1/SEM1 complexes forming structures that mediate binding to single or double stranded DNA. The binding of DSS1 the DBD region of BRCA2 DBD is documented to have a vital role in sustaining its solubility and stability enabling it to perform its usual functions of DNA break repair and consequent tumor suppression by interacting with other partner genes. For example, The SEM1/DBD complex is reported to be involved in the nuclear transport of BRCA2/RAD51 complexes followed by loading of RAD51 on RPA coated single stranded DNA thereby initiating Homology Directed Repair (HDR). RAD51 and its gene family are known to be critically involved in regulating the fidelity of DNA through playing varied roles in molecular cellular processes including: double strand break repair, replication stress, and meiosis. BRCA2 mutations involving BRC2/SEM1 interaction region may thus cost the gene loss of its interaction with SEM1 and lead to the development of different tumors as a result of its inability to interact with this gene in performing its DNA repair and stabilization functions. The mediator role of BRCA2 in homologous recombination has been documented in Le, et al. The specific mutation explained above may impair this role by making BRCA2 unable to interact well with SEM1/DSS1.
FA Complementation group D2 (FANCD2) gene codes for FA Complementation group D2 (FANCD2) protein, an FA (Fanconi Anemia) protein which is involved in repairing of damaged DNA. FANCD2 and other FA proteins function together to repair damaged DNA by activating the Fancony Anemia (FA) pathway the primary step of which is the mono-ubiquitination of the FANCD2 and FANCI proteins. This mono-ubiquitination step occurs upon exposure of the proteins to DNA damaging agents and during the s phase of the cell cycle. Inactivation of the FA pathway way leads to fanconi anemia, a rare genetic disease characterized by increased risk for bone marrow failure and cancer. Fanconi anemia phenotypes are generally characterized by chromosomal instability and hypersensitivity to natural or artificial DNA Interstrand Cross Linking (ICL) agents. FA proteins play their functional roles in the cell by sensing, recognizing and processing of ICL agents. A direct interaction between FANCD2 and BRCA2 proteins has been documented which has been mapped to part of the highly conserved BLAT domain in BRCA2.
It is through this interaction that DNA interstrand cross links are repaired FANCD2 recruites BRCA2 in to Interstrand Crosslinks (ICL) for repair after sensing them [23]. BRA2 mutation in the region where it interacts to FABCD2 can, therefore, lead to its inability to be recruited to chromosomal regions of DNA interstrand crosslinks for repair. The two SNPs: rs80358974 (substitution of isoleucine by threonine) and rs80358730 (substitution of glycine by serine) may thus be associated with the occurrence of fanconi anemia.
The 7 SNPs chosen for analysis were predicted to be found in 100% conserved residue by hope web based server. Evolutionarily conserved residues harbor amino acids participating in important biological processes. These regions may be enzyme active sites or regions of active protein protein interactions. Mutations occurring at these sites are thought to be more deleterious compared to those which occur at non conserved positions. Our identified and predicted SNPs, may, therefore, be involved in the development of different human clinical conditions. Finally the predicted hydrophobicity and charge difference between the wild type proteins may affect the ionic and hydrophobic interactions of the protein leading to altered cellular functions.
BRCA2 gene is involved in many cellular processes its major function being DNA break repair by homologous recombination. BRCA2 interacts with various genes such as RAD51, PALB2, FANCD2, SEM1 etc. in performing its functions. Non synonymous deleterious mutations can happen to this gene in the form of single nucleotide polymorphisms a significant number of which can be deleterious altering the structure and function of the protein. BRCA2 SNPs reported in NCBI ClinVar as variants of unknown significance and variants with conflicting interpretations of pathogenicity can be analyzed by in silico methods to predict their clinical significance. Therefore, this study analyzed BRCA2 variants with conflicting interpretations of pathogenicity from NCBI by using bioinformatics tools and has found deleterious SNPs. Some of these SNPs were found to harbor biologically important mutations which may be associated to different clinical conditions.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Zenebe B, Nigussie H, Belay G, Seboka N (2023) In silico Analysis of Non-Synonymous Coding Single Nucleotide Polymorphisms (SNPs) in Human BRCA2 Gene. J Proteomics Bioinform. 16:641.
Received: 13-Dec-2022, Manuscript No. JPB-22-20855 ; Editor assigned: 16-Dec-2022, Pre QC No. JPB-22-20855 (Q); Reviewed: 30-Dec-2022, QC No. JPB-22-20855 ; Revised: 17-Mar-2023, Manuscript No. JPB-22-20855 (R); Published: 24-Mar-2023 , DOI: 10.35248/0974-276X.23.16.641
Copyright: © 2023 Zenebe B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.