
Journal of Clinical and Cellular Immunology
Open Access
ISSN: 2155-9899

ISSN: 2155-9899
Research Article - (2025)Volume 16, Issue 1
Aim: In search of an effective treatment for COVID-19 disease, in Bangladesh, a group of doctors reported surprising success with two commonly used drugs, Ivermectin and Doxycycline. But the mechanism underlying therapeutic action of these drugs remains obscure. The current study explores the possible molecular targets and mechanism of action.
Methods: Structure of both Ivermectin and Doxycycline were optimized using universal forcefield as well as by quantum mechanics. Molecular docking and molecular dynamics simulation techniques were employed to explore the possible mechanism of action of these drugs. Effectiveness of Ivermectin and Doxycycline was evaluated against several proteins of SARS-CoV-2 and human ACE2 receptor.
Results: It was found that both the drugs have significant binding affinity with SARS-CoV-2 proteins as evinced from low predicted binding energy. Ivermectin showed better binding than Doxycycline. Ivermectin showed a perfect binding to the site of interaction between Spike-RBD and ACE2 proteins indicating that it might be interfering with the viral entry in to the host cells. It also exhibited significant binding affinity with different SARSCoV- 2 structural and non-structural proteins which have diverse functions in virus life cycle. Our study also shows that Ivermectin has lower binding affinity to spike protein of omicron variant compared with wild-type. Significant binding of Ivermectin with RdRp indicated its role in the inhibition of the viral replication and ultimately impeding the multiplication of the virus. Ivermectin also showed significant binding affinity with the proteins which help the virus in escaping from host immune system. Molecular dynamics study well corroborates with molecular docking and showed that binding of Ivermectin with Mpro, Spike, NSP3, NSP16 and ACE2 are stable.
Significance: This study predicts that the combination of Ivermectin and Doxycycline might be executing the therapeutic effect by inhibition of viral entry, moderation of viral multiplication and improving the viral clearance.
COVID-19; SARS-CoV-2; Ivermectin; Doxycycline; RNA dependent RNA polymerase; Molecular docking; Molecular dynamics simulation
The ongoing outbreak of the Coronavirus disease 2019 (COVID-19) has affected almost entire World and it has become a pandemic. Because of high rate of transmission and infectivity of SARS-CoV-2, World Health Organization (WHO) had declared it as a global disaster. In a span of >36 months, COVID-19 disease has caused over 681 million human infections and about 6.8 million fatalities. Like other CoVs, SARS-CoV-2 has a positive-sense single-stranded RNA genome and four structural proteins, namely the Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) proteins. It uses the surface spike protein and interacts with human Angiotensin- Converting Enzyme 2 (ACE2) receptor present on the host cell surface. Unlike, other CoVs, SARS-CoV-2 spike protein has high affinity to the human ACE2 receptor [1].
Knowledge gained since 2019, shows that there is rapid spreading of SARS-CoV-2 Variants of Concern (VoCs). This demonstrates how easily this virus can accommodate antigenic changes in its spike protein without any loss of it properties. Until now several variants of the SARS-CoV-2 has been identified with different level of the severity and infectivity. The current VoC, omicron has lower severity but is of major concern because of the high infectivity rate compared with previous variants such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) VoCs. It has a peculiar spectrum of mutations characterized by the acquisition of mutations or deletions rarely detected in previously identified variants, particularly in the spike glycoprotein [2,3].
Currently a lot of studies are going on to find possible therapeutics to manage the COVID-19 disease. Until now, nonpharmaceutical interventions like social distancing, isolation of the infected individual and the quarantine of the exposed individuals have resulted in improved management of the COVID-19 crisis. Most of the current research for development of effective therapeutics against SARS-CoV-2 is based on the screening of existing broad-spectrum anti-virals, small molecule libraries, FDA approved drug libraries and traditional alternative medicines [4-13]. Until now several vaccines have come-up with limited usage and many more are at the final stage of their release [14-16]. Accumulating evidence suggests that even double-dose-vaccinated and convalescent people are not well protected against the omicron variant [17,18]. Apart from this, many of the new chemical entities and re-purposed drugs are still being tested against SARS-CoV-2 [19-21].
In the month of May, 2020, a team of the doctors from Bangladesh Medical College Hospital, reported that Ivermectin given as a single dose with doxycycline, yielded significant results in curing COVID-19 patients. This finding attracted several researchers all over the globe to understand and explore the possible mechanism of the action for this combination against COVID-19 [22,23]. Until now several report related to beneficial effect of Ivermectin has been reviewed [24,25]. Apart from this several clinical trials were also started with either Ivermectin alone or in combination with other drugs which were recently reviewed by Kaur, et al. 2021 [26]. Some of the clinical trials such as NCT04591600, NCT04523831, NCT04407130, and NCT04551755 were based on the combination of Ivermectin and Doxycycline. Out of these clinical trials only result for NCT04591600 and NCT04523831 are available. In NCT04591600 clinical trial, a total 70 COVID-19 patients consisting of 48 mild to moderate, 11 severe, and 11 critical patients were recruited and it was observed that Ivermectin and Doxycycline treatment reduces the time of recovery and also decreases the percentage of patients who progress to more advanced stage of disease. This treatment also reduces the mortality rate in severe patients from 22.72% to 0%, however 18.2% of critically ill patients died. From this study it was also learnt that early administration of Ivermectin with Doxycycline will have higher success rate [27]. In clinical trial NCT04523831, it was observed that a number of patients had early clinical improvement in term of body temperature, respiratory symptoms, lung imaging, no co-morbidities or complications within 7 days. Participants with late clinical recovery required more than 12 days for clinical improvement as defined above. It was also found that number of patients who remained RT-PCR positive for COVID-19 at day 14 also reduced.
Ivermectin is a FDA-approved broad spectrum anti-parasitic drug [28]. Safety, tolerability, and pharmacokinetics of Ivermectin is well established and it is well tolerated, with no indication of central nervous system toxicity at doses up to 10 times the higher than the FDA‐approved dose [29]. It affects a plethora of invertebrate species, including parasitic arachnids, nematodes and insects. It has been earlier shown to exert anti-viral activity in vitro against Dengue fever virus (DENV), influenza virus, West Nile Virus and Venezuelan equine encephalitis virus [30-32]. Very recently, Caly, et al. have shown the efficacy of Ivermectin in Vero-hSLAM cells infection with SARS-CoV-2 and observed an effective loss of viral material within 48 h. Based on their finding, Caly, et al. hypothesised that it might we working by inhibiting importin α/β1 (IMPα/β1)-mediated nuclear import of viral proteins, as shown for other RNA viruses [33-35]. Ivermectin has a very well established safety profile for human use [36]. The second drug, doxycycline is a synthetic antibiotic derived from tetracycline and have broad spectrum antibiotic property. Doxycycline other than antibiotics, are known to inhibit M2-type polarization of macrophages [37]. Doxycycline is a FDA approved antibiotic since 1967 and is extensively referred by most clinicians. Similar to Ivermectin, Doxycycline also has a long history of safety in humans. Different groups of researcher have tried to explore the possible mechanism of action of Ivermectin against SARS-CoV-2 using in silico approach. But no one has tried to explore how the combination of Ivermectin with Doxycycline might be working synergistically to get the positive response in the COVID-19 patients. In a recent study a comparison of effectiveness of the Ivermectin was studies in a range of SARS-CoV-2 variants and it was found that it has a relatively homogeneous in vitro activity against different variants [38].
In the modern era, application of the molecular docking [39] and Molecular Dynamics (MD) simulation [40] have become the integral part to fasten the drug development. Molecular docking provides information about docking scores based on the binding energies whereas MD simulation bridges the theoretical study with the experimental study. Thus both these tools have enhanced the drug screening process before going to test them in the animal models. In the current study, I have explored the possible mechanisms to explain the above experimental and clinical findings using molecular docking and MD simulation studies. For this purpose, Ivermectin and Doxycycline were docked with different viral proteins such as Spike, Main Protease (Mpro), Papain-like protease (PLpro), RNA dependent RNA Polymerase (RdRp), Nucleocapsid (N-protein), and several Non- Structural Proteins (NSPs) like NSP3, NSP10, NSP15 and NSP16, and human Angiotensin Converting Enzyme 2 (ACE2) receptor. Our molecular docking study predicted that Ivermectin and Doxycycline both have significant binding affinity for SARSCoV- 2 spike and ACE2 interacting region, indicating poor interaction for viral entry inside the cells. The predicted binding energy of Ivermectin is much better than that of Doxycycline. At the same time, Ivermectin has significantly high binding affinity with the RdRp (also known as NSP12), indicating poor viral genome replication for viral multiplication. Ivermectin significantly inhibited ADP Ribose Phosphatase (NSP3), Endoribonuclease (NSP15) and methyltransferase (NSP10- NSP16 complex) of SARS-CoV-2 which were involved in the virus escape from the host innate immune system. Ivermectin and Doxycycline both bind with SARS-CoV-2 Mpro adjacent to its active site.
Thus, the major objective of the current study is to explore the mechanism underlying therapeutic action of Ivermectin and Doxycycline combination towards SARS-CoV-2. So we have explores the interaction of both drugs with different possible molecular targets and predicted their mechanism of action.
Chemical structure of molecules and target proteins: Chemical structures information for Ivermectin (PubChem CID-6321424) and doxycycline (PubChem CID-54671203) were downloaded from PubChem. Both the molecules were energy minimized Using Universal Forcefield (UFF) before proceeding for molecular docking. In order to study the mode of interaction of Ivermectin and Doxycycline with various SARS-CoV-2 proteins and receptors found on host cells, following PDB ID’s 6lu7 (Mpro), 6m0j (spike, wild-type), 7wlc (spike of omicron), 6m71 (RdRp, NSP12), 6w02 (NSP3), 6w9c (PLpro), 6m3m (Nucleocapsid), 6w4b (NSP9), 6wkq (NSP10), 6vww (NSP15), 6wkq (NSP16) and 6m0j (ACE2) were used. All the protein structures were retrieved from protein data bank and cleaned using USCF Chimera software [41].
Molecular docking: PyRx virtual screening tool was used for preparation of the ligands and receptors to perform the molecular docking. Molecular docking was performing using Vina wizard of PyRx virtual screening tool [42,43]. For preparation of protein input files, all the water molecules, ligands and ions were removed from *.pdb files and polar hydrogens were added before saving *.pdbqt format. Both the molecules were energy minimized and saved in *.pdbqt format after adding polar hydrogens for further docking process. Following AutoDock Vina docking parameters such as (center_x=-26.2834, center_y=12.5988, center_z=58.9648, size_x=66.124, size_y=22.9423, size_z=51.456), (center_x=-32.483, center_y=26.077, center_z=7.923, size_x=52.974, size_y=46.699, size_z=30.699) and (center_x=-25.0997, center_y=19.903, center_z=3.047, size_x=78.872, size_y=67.603, size_z=35.866) were used for SARS-CoV-2 Mpro (PDB ID: 6LU7), SARS-CoV-2 spike (PDB ID: 6m0j) and human ACE2 (PDB ID: 6m0j) respectively. For docking RdRp (PDB ID: 6m71) NSP7 and NSP8 was removed from the NSP12 and grid box for docking parameter was set at the NSP12-NSP7 and NSP12-NSP8 interface. Docking parameters used for NSP12 were center_x=112.289, center_y=131.245, center_z=141.597, size_x=83.597, size_y=69.959, size_z=45.240. A blind docking was performed for the remaining target receptor proteins.
Structure optimization: Structure of both Ivermectin and Doxycycline were also optimized using DFT employing ORCA 5.0, an ab initio quantum chemistry program package [44]. We have used Hartee-Fock-3c (HF-3c) method because it is most widely used method for optimization of the geometries of small organic molecules [45]. Binding energies of these optimized structures with different target proteins were also compared with the binding energies obtained from structure optimized using UFF.
Interactions of the ligand with critical residues: 2D and 3D interactions of docked complex were analysed for the visualization of the best binding poses. As the importance of hydrogen bonding is well documented in drug discovery and design, further hydrogen bond and other non-polar interactions of best binding poses were examined using Discovery studio visualizer software.
Molecular dynamics simulation: The protein-ligand complex for spike, Mpro, NSP3, NSP16 and human ACE2 receptor with Ivermectin were obtained after the molecular docking study. The simulation systems for protein-ligand complex with or without Ivermectin were performed using the GROMACS (2019.4) software [46]. Protein topology was prepared using pdb2gmx module of GROMACS. Whereas ligand topology was derived from Prodrg server [47]. MD simulation was performed with Gromos53a6.ff force-field using the GROMACS. Proteins were solvated using SPC (Simple Point Charge) water model and counter ions were added to neutralize the system. After that, system was energy minimized and further water and ions were allowed to equilibrate around the protein in a two-step equilibration process. The first part of equilibration was at a constant Number of particles, Volume, and Temperature (NVT). The second part of equilibration was at a constant Number of particles, Pressure, and Temperature (NPT). NVT and NPT equilibration simulations were performed for 100 ps. Following equilibration, the production simulation duration was performed for 250 nanoseconds (ns). The trajectory data were saved at different time points to analyse the change in the dynamics of protein-ligand complex. MD simulation data were also processed for finding several other information for the protein-ligand interaction.
Trajectory analysis: Trajectory analysis was performed using inbuilt GROMACS analysis tools. After completion of MD simulation, results for stability and flexibility for protein-ligand complexes were analysed using Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) using gmx rms, and gmx rmsf tools, respectively. Further, H-bond interaction between different SARS-CoV-2 proteins and Ivermectin was analysed using gmx hbond tools selecting protein and ligand. The Principal Component Analysis (PCA) has been extensively used to analyse and investigate the global motion of a protein during the simulation. It can convert the highdimensional data of protein dynamics into the low-dimensional space to obtain a series of eigenvectors and eigenvalues that reflect overall motions in the protein. PCA was obtained by diagonalizing and solving the eigenvalues and eigenvectors for the covariance matrix. The covariance matrix for illustration of PCA was calculated using GROMACS analysis tool gmx cover which builds and diagonalizes the covariance matrix. Another GROMACS analysis tool, gmx anaeig was utilized to calculate the overlap between principal components and coordinates of the trajectory.
MM/PBSA binding free energy: Docking of molecule with a protein at their binding site is a quick method to find the probable binding pose of a compound. However, this methodology still lacks the capacity to appropriately rank the affinities of the molecule to the target protein. Prediction of the binding energies of a lead molecule with biological targets plays an important role in structure-based drug design. Thus in order to have an enhanced ranking of the ligands and determination of their predictive binding energies, Mechanic/Poisson- Boltzmann Surface Area (MM/PBSA) calculations were implemented with MD generated output. The binding free energy provides an overview of the bio-molecular interactions between protein and ligand which constitutes of potential energy, polar and non-polar solvation energies. A more negative binding free energy values indicate a stronger binding. For calculation of MM/PBSA binding free energies ‘g_mmpbsa’ script of GROMACS was used [48]. The binding energy calculations in this method were done using the following equation:
deltaGbinding=Gcomplex-(Greceptor-Gligand)
The deltaGbinding represents the total binding energy of the complex, while the binding energy of free receptor is Greceptor, and that of unbounded ligand is represented by Gligand.
Steered molecular dynamics simulations: Interactions between proteins and ligands play an important role in many biological processes. For comparing the Ivermectin interaction with wild type and omicron variant spike protein, steered MD simulation (SMD) was employe. Basically, SMD is a kind of advance MD simulation which mimics Atomic Force Microscopy (AFM). It is very good computational approach for the examination of detachment of a ligand from its protein complex so it resembles with the AFM experiment. In the current study, SMD was performed by applying a time-dependent external force on Ivermectin to facilitate its unbinding from wild-type and omicron variant spike proteins, which cannot usually be achieved by conventional MD simulation. During the SMD simulations, the force was only applied along the pulling direction and velocity was kept constant. The spring constant was set to the value of 1000 kJ mol-1 nm-2 and the pulling velocity was 0.001 nm per pico second (10 A/ns). The time length for each simulation was set to 200 ps to ensure the complete unbinding of Ivermectin from Spike.
Molecular docking: Molecular docking study revealed that both Ivermectin and Doxycycline have significant binding affinity with various SARS-CoV-2 proteins i.e., Mpro, spike, PLpro, RdRp, nucleocapsid, NSP3, NSP9, NSP10, NSP15, NSP16 and host ACE2 receptor. The more the negative value of the binding energy, the better will be binding affinity of the molecule with it target protein. Ivermectin showed better binding affinity in term of the predicted binding energy with the target proteins compared to doxycycline. From the Figure 1A, it is clear that Ivermectin independently bind at the interacting region of viral spike and host ACE2 receptor indicating that it might hamper the interaction of spike and ACE2 receptor and inhibit the virus entry inside the cells. Figure 1B, shows the 2D interaction of Ivermectin and doxycycline with different protein residues of spike and ACE2. The predicted binding energies of Ivermectin and doxycycline are -8.7 kcal/mol and -7.0 kcal/mol for spike protein of wild-type and -7.5 kcal/mol and -7.6 kcal/mol for ACE2 receptor respectively. The predicted binding energies of Ivermectin and doxycycline are -7.7 kcal/mol and -6.4 kcal/mol for spike protein of omicron which is lower than the wild-type variant. Figure 1C, shows the 3D interaction of Ivermectin with the SARS-CoV-2 spike and human ACE2 receptor. As shown in the figure 1C, Ivermectin has conventional hydrogen bonding with residues GLN 409 and TYR 453 as well as alkyl or pi-alkyl binding with residues LYS 417 and LEU 455 of SARS-CoV-2 spike protein. From figure 1C, it is also clear that Ivermectin has conventional hydrogen bonding with residues ASN 33, GLN 383 and ARG 393, carbon-hydrogen bonding with residues GLU 37 and ALA 287, and Alkyl or pi-alkyl binding with residues LYS 26, LEU 29, HIS 34, VAL 93, PRO 389 and LEU 392 of the human ACE2 receptor protein (Tables 1 and 2).
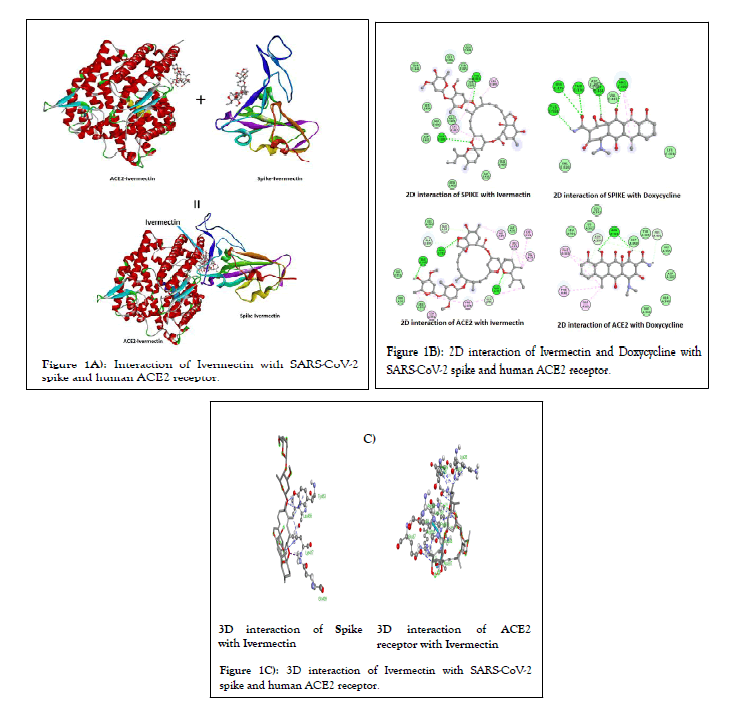
Figure 1: Interaction of Ivermectin with SARS-CoV-2 spike and human ACE2 receptor.
| Targets | Pose | Predicted binding energy (kcal/mol) | Predicted inhibition constant, Ki (µM) |
|---|---|---|---|
| Mpro, 6lu7 | Pose 1 | -8.4 | 0.69 |
| Pose 2 | -8.3 | 0.81 | |
| Pose 3 | -8 | 1.35 | |
| PLpro, 6w9c | Pose 1 | -8.5 | 0.58 |
| Pose 2 | -8.2 | 0.96 | |
| Pose 3 | -7.6 | 2.65 | |
| Spike, 6m0j (Wild type) | Pose 1 | -8.7 | 0.41 |
| Pose 2 | -8.3 | 0.81 | |
| Pose 3 | -8.2 | 0.96 | |
| Spike, 7wlc (Omicron) | Pose 1 | -7.7 | 2.24 |
| Pose 2 | -7.6 | 2.65 | |
| Pose 3 | -7.6 | 2.65 | |
| Nucleocapsid, 6 m 3 m | Pose 1 | -8.2 | 0.96 |
| Pose 2 | -8.1 | 1.14 | |
| Pose 3 | -7.9 | 1.6 | |
| nsp3, 6w02 | Pose 1 | -8 | 1.35 |
| Pose 2 | -7.8 | 1.89 | |
| Pose 3 | -7.2 | 5.21 | |
| nsp9, 6w4b | Pose 1 | -7.9 | 1.6 |
| Pose 2 | -7.5 | 3.14 | |
| Pose 3 | -7.3 | 4.4 | |
| nsp10, 6wkq | Pose 1 | -8.9 | 0.29 |
| Pose 2 | -8.3 | 0.81 | |
| Pose 3 | -8.2 | 0.96 | |
| nsp12, 6m71 | Pose 1 | -9.1 | 0.21 |
| Pose 2 | -8.9 | 0.29 | |
| Pose 3 | -8.8 | 0.35 | |
| nsp15, 6vww | Pose 1 | -8.4 | 0.69 |
| Pose 2 | -8.3 | 0.81 | |
| Pose 3 | -7.9 | 1.6 | |
| nsp16, 6wkq | Pose 1 | -8.3 | 0.81 |
| Pose 2 | -7.8 | 1.89 | |
| Pose 3 | -7.5 | 3.14 | |
| ACE2, 6m0j | Pose 1 | -7.5 | 3.14 |
| Pose 2 | -7.4 | 3.71 | |
| Pose 3 | -7.2 | 5.21 |
Table 1: Predicted binding energy and inhibition constant of Ivermectin for various SARS-CoV-2 target proteins and human ACE2 receptor.
| Targets | Pose | Predicted binding energy (kcal/mol) | Predicted inhibition constant, Ki (µM) |
|---|---|---|---|
| Mpro, 6lu7 | Pose 1 | -7.1 | 6.16 |
| Pose 2 | -6.6 | 14.35 | |
| Pose 3 | -6.4 | 20.12 | |
| PLpro, 6w9c | Pose 1 | -7.1 | 6.16 |
| Pose 2 | -6.5 | 16.99 | |
| Pose 3 | -6.3 | 23.82 | |
| Spike, 6m0j (Wild type) | Pose 1 | -7 | 7.3 |
| Pose 2 | -6.7 | 12.12 | |
| Pose 3 | -6.5 | 16.99 | |
| Spike, 7wlc (Omicron) | Pose 1 | -6.4 | 20.12 |
| Pose 2 | -6.4 | 20.12 | |
| Pose 3 | -6.3 | 23.82 | |
| Nucleocapsid, 6m3m | Pose 1 | -6.3 | 23.82 |
| Pose 2 | -6.2 | 23.82 | |
| Pose 3 | -6.1 | 33.4 | |
| nsp3, 6w02 | Pose 1 | -7.1 | 6.16 |
| Pose 2 | -6.4 | 20.12 | |
| Pose 3 | -6.3 | 23.82 | |
| nsp9, 6w4b | Pose 1 | -6.1 | 33.4 |
| Pose 2 | -6 | 39.54 | |
| Pose 3 | -5.8 | 55.44 | |
| nsp10, 6wkq | Pose 1 | -6.9 | 8.64 |
| Pose 2 | -6.8 | 10.23 | |
| Pose 3 | -6.7 | 12.12 | |
| nsp12, 6m71 | Pose 1 | -7.9 | 1.6 |
| Pose 2 | -7.4 | 3.71 | |
| Pose 3 | -7.3 | 4.4 | |
| nsp15, 6vww | Pose 1 | -7.5 | 3.14 |
| Pose 2 | -7.2 | 5.21 | |
| Pose 3 | -6.8 | 10.23 | |
| nsp16, 6wkq | Pose 1 | -7.7 | 2.24 |
| Pose 2 | -7.5 | 3.14 | |
| Pose 3 | -7.4 | 3.71 | |
| ACE2, 6m0j | Pose 1 | -7.6 | 2.65 |
| Pose 2 | -6.6 | 14.35 | |
| Pose 3 | -6.6 | 14.35 |
Table 2: Predicted binding energy and inhibition constant of Doxycycline for various SARS-CoV-2 target proteins and human ACE2 receptor.
Figures 2A and 2B, shows the predicted binding site and 2D interaction of Ivermectin and doxycycline for SARS-CoV-2 RdRp. The anticipated binding energies of Ivermectin and doxycycline for RdRp are -9.1 and -7.9 kcal/mol respectively. Figures 3A and 3B, shows the predicted binding site and 2D interaction of Ivermectin and doxycycline with SARS-CoV-2 NSP3 and PLpro. The predicted binding energies of Ivermectin and Doxycycline -8.0 and -7.1 kcal/mol for NSP3 whereas -8.5 and -7.1 kcal/mol for PLpro. Figures 4A and 4B, shows the predicted binding site and 2D interaction of Ivermectin and doxycycline with SARS-CoV-2 NSP16 and NSP10. Our study shows that Ivermectin binds to the S-Adenosylmethionine binding site of NSP16 with a binding affinity evinced from low predicted binding energy (-8.3 kcal/mol). It also binds with the NSP16 cognate activation partners NSP10 with a binding energy of -8.9 kcal/mol.
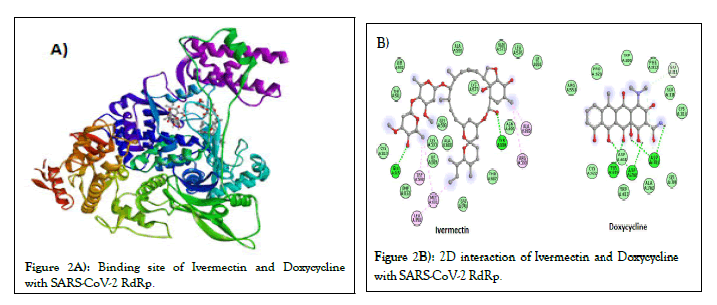
Figure 2: Binding site of Ivermectin and Doxycycline with SARS-CoV-2 RdRp.
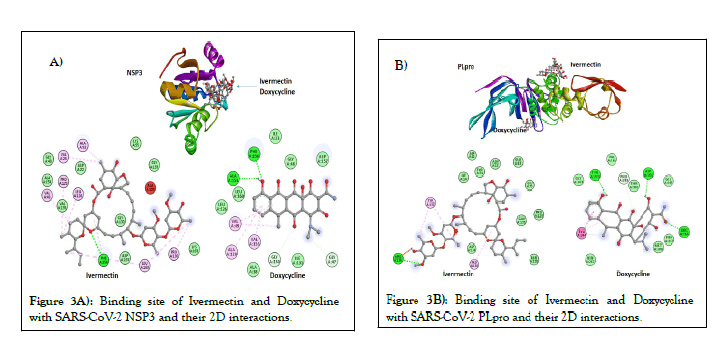
Figure 3: Binding site of Ivermectin and Doxycycline with SARS-CoV-2 NSP3 and their 2D interactions.
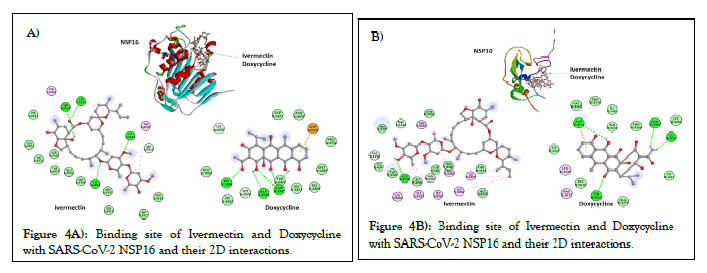
Figure 4: Binding site of Ivermectin and Doxycycline with SARS-CoV-2 NSP16 and their 2D interactions.
Some of the other important CoVs proteins such as Nucleocapsid (N), NSP9 and NSP15 were also used for molecular docking study. The predicted binding energies of Ivermectin and doxycycline for N-protein are -8.2 kcal/mol and -6.3 kcal/mol respectively. The predicted binding energies of Ivermectin and doxycycline are -7.9 kcal/mol and -6.1 kcal/mol for NSP9 whereas -8.4 kcal/mol and -7.5 kcal/mol for NSP15. All together these finding show that Ivermectin have better binding affinity with N-protein, NSP9, and NSP15 compared to doxycycline.
All the above binding energies were predicted with the ligand structure optimized using UFF. When molecular docking was performed using DFT optimized structure a slight variation at few places in energies were predicted (Table 3).
| Drugs | Targets | Predicted binding energy (kcal/mol) | Predicted Inhibition constant, Ki (µM) |
|---|---|---|---|
| Ivermectin | Mpro, 6lu7 | -8.4 | 0.685366 |
| PLpro, 6w9c | -8.5 | 0.578816 | |
| Spike, 6m0j (Wild type) | -7.8 | 1.88 | |
| Spike, 7wlc (Omicron) | -5 | 214 | |
| Nucleocapsid, 6m3m | -7.1 | 6.16 | |
| nsp3, 6w02 | -8.1 | 1.13 | |
| nsp9, 6w4b | -7.9 | 1.595262 | |
| nsp10, 6wkq | -9 | 0.24 | |
| nsp12, 6m71 | -8.5 | 0.57 | |
| nsp15, 6vww | -9.3 | 0.15 | |
| nsp16, 6wkq | -9.3 | 0.15 | |
| ACE2, 6m0j | -9.3 | 0.15 | |
| Doxycycline | Mpro, 6lu7 | -7.3 | 4.39 |
| PLpro, 6w9c | -8.2 | 0.96 | |
| Spike, 6m0j (Wild type) | -7.7 | 2.23 | |
| Spike, 7wlc (Omicron) | -5.9 | 46.8 | |
| Nucleocapsid, 6m3m | -6.7 | 12.11 | |
| nsp3, 6w02 | -6.8 | 10.2 | |
| nsp9, 6w4b | -6.7 | 12 | |
| nsp10, 6wkq | -7.4 | 3.7 | |
| nsp12, 6m71 | -8.8 | 0.35 | |
| nsp15, 6vww | -8.7 | 0.413 | |
| nsp16, 6wkq | -8.4 | 0.68 | |
| ACE2, 6m0j | -8.6 | 0.488 |
Table 3: Predicted binding energy and inhibition constant of Ivermectin and Doxycycline which structures was optimized using quantum mechanics for various SARS-CoV-2 target proteins and human ACE2 receptor
MD simulation and stability of Ivermectin with target proteins: MD simulations were carried out for SARS-CoV-2 spike, Mpro, NSP3, NSP16 and human ACE2 receptor and Ivermectin complex in the solvated states for 250 ns. The results of MD simulations have been examined on the basis of Root Mean Square Deviation (RMSD) by selection of protein and Root Mean Square Fluctuation (RMSF) using C-α. RMSD is a well-known estimator of equilibration and protein stability. Based on the obtained MD trajectories, RMSD values of the protein backbone in complexes with Ivermectin were computed. The RMSD plot analysis (average of three MD productions) indicates that for all the target proteins, the RMSD values showed an initial steady increase and then reached equilibrium during the rest simulation period (Figure 5). However, any variations noticed were in order of 0.15 nm showing that the complex has not undergone large conformational changes (Figure 5). To explore the local protein flexibility, the time average of RMSF values of spike, Mpro, NSP3, NSP16 and ACE2 protein in presence Ivermectin over simulation period were calculated. RMSF with respect to amino acid for all the studied proteins were plotted and we did not find any major fluctuation except for a few places where moderate fluctuations were observed (Figure 6).
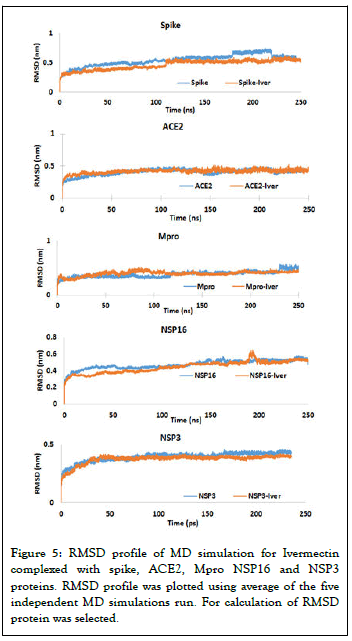
Figure 5: RMSD profile of MD simulation for Ivermectin complexed with spike, ACE2, Mpro NSP16 and NSP3 proteins. RMSD profile was plotted using average of the five independent MD simulations run. For calculation of RMSD protein was selected.
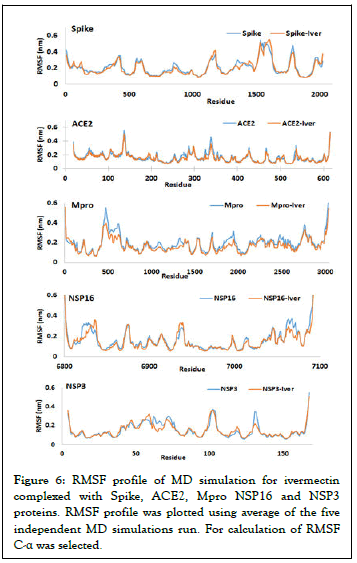
Figure 6: RMSF profile of MD simulation for ivermectin complexed with Spike, ACE2, Mpro NSP16 and NSP3 proteins. RMSF profile was plotted using average of the five independent MD simulations run. For calculation of RMSF C-α was selected.
Hydrogen bond analysis: Non-covalent interactions like hydrogen bonds (H-bonds) are key-players in the formation of a protein-ligand complex. In H-bonding, attractive potential results from a partially positively charged hydrogen pointing to a partially negatively charged acceptor atom, e.g. oxygen or nitrogen. The analysis of H-bonds can give valuable insights about the stability of a ligand-protein complex. More detailed analysis was performed at atomic level by calculation of the number of hydrogen bonds formed in all the SARS-CoV-2 target proteins with Ivermectin. Figure 7 shows inter molecular Hbonding in Spike, Mpro, NSP3 and NSP16 with the Ivermectin in their complex. Our study shows that there was an average Hbond was 2.67, 0.9, 0.8, and 1.56 for Spike, Mpro, NSP3 and NSP16 with Ivermectin respectively.
Principal component analysis: PCA of molecular dynamics trajectories of SARS-CoV-2 spike, Mpro, NSP3, NSP16 and human ACE2 was conducted to study the collective motion of the respective unbound and Ivermectin-bound protein on the trajectories generated by MD simulations. The overlap between principal components and coordinates of the trajectory are shown in Figure 8. During PCA, lower eigenvalues signify lower expansion of protein indicating more compactness throughout the simulation. Figure 8 shows that Ivermectin bound proteins covered a smaller region of phase space compared with the unbound proteins for all the studied complexes.
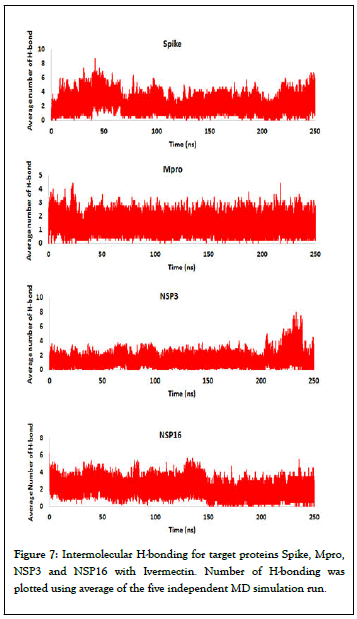
Figure 7: Intermolecular H-bonding for target proteins Spike, Mpro, NSP3 and NSP16 with Ivermectin. Number of H-bonding was plotted using average of the five independent MD simulation run.
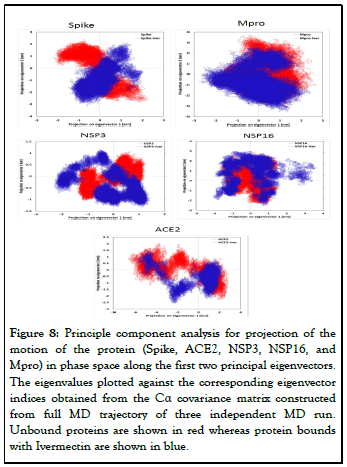
Figure 8: Principle component analysis for projection of the motion of the protein (Spike, ACE2, NSP3, NSP16, and Mpro) in phase space along the first two principal eigenvectors. The eigenvalues plotted against the corresponding eigenvector indices obtained from the Cα covariance matrix constructed from full MD trajectory of three independent MD run. Unbound proteins are shown in red whereas protein bounds with Ivermectin are shown in blue.
MM/PBSA calculation: After performing simulation, MD output file were further used to calculate average free binding energy using g_mmpbsa package and MmPbSaStat.py python script (Table 4). The final binding energy is a cumulative sum of van der Wall, electrostatic, polar solvation, and SASA energy. As shown in Table 4, except for the polar solvation energy, all other forms of energy contributed favourably to the interaction between different targets with Ivermectin. The calculated MM/PBSA free binding energy of Ivermectin are -23.97, -106.78, -91.07, -65.53, and -46.17 kJ/mol for Spike, Mpro, NSP3, NSP16, and ACE2 respectively. Binding energy obtained from MM/PBSA calculation corroborated positively with the findings of molecular docking study.
| Energies | kJ/mol | ||||
|---|---|---|---|---|---|
| Spike ± S.E.M | Mpro ± S.E.M | NSP3 ± S.E.M | NSP16 ± S.E.M | ACE2 ± S.E.M | |
| van der Waal energy | -68.94 ± 2.58 | -154.73 ± 28.8 | -128.37 ± 10.07 | -120.04 ± 12.69 | -68.71 ± 13.93 |
| Electrostatic energy | -7.66 ± 2.46 | -12.89 ± 4.31 | -23.46 ± 11.05 | -23.92 ± 9.29 | -6.95 ± 16.27 |
| Polar solvation energy | 27.31 ± 6.40 | 77.06 ± 12.65 | 74.85 ± 15.44 | 90.97 ± 11.45 | 56.57 ± 24.12 |
| SASA energy | -7.47 ± 0.32 | -16.22 ± 2.86 | -14.09 ± 1.32 | -12.53 ± 1.68 | -6.18 ± 1.597 |
| Binding energy | -56.76 ± 2.52 | -106.78 ± 23.69 | -91.07 ± 7.12 | -65.53 ± 15.88 | -25.261± 6.05 |
Table 4: MM/PBSA free binding energy of Ivermectin for various SARS-CoV-2 target proteins and human ACE2 receptor.
Further, we evaluated the contribution of each residue of Spike, NSP3, NSP16, Mpro, and ACE2 in terms of binding free energy with Ivermectin. Figure 9 shows the contribution of each residue energy calculated by decomposing the total binding free energy of the system. Our study shows that the residues PHE374, TRP436, LEU441, VAL445, PRO499, VAL 503, and TYR 505 of Spike are major contributor towards binding with Ivermectin. For NSP3, amino acid residue ILE23, VAL24, ALA38, VAL49, ILE131, PHE132, and PHE156 are the major contributor towards Ivermectin binding. For NSP16, residue CYS6823, LEU6825, TYR6835, TYR6930, VAL6938, THR6970, SER6999, THR7040, and PHE7043 have major contribution towards binding. For Mpro, amino acid residue LEU208, TRP218, LEU232, MET235, TYR237, PRO241, LEU271, LEU272, MET276, LEU281, and LEU286 are the major contributor towards binding with Ivermectin. As for contribution of different amino acid residue of ACE2 is concerns, THR324, GLN325, TRP328, ASP350, ASP355, and MET383 play major contribution towards binding with Ivermectin.
Steered molecular dynamics simulations: During SMD simulation, an external force is applied to the Ivermectin to pull it away from the spike protein. Figure 10 shows the rupture forces for the Ivermectin-spike complex plotted in respect to time along with Ivermectin moving away from the binding site. The pulling simulation shows that lower rapture force is required to remove the Ivermectin from the omicron variant spike protein compared to wild-type variant. The rapture force to unwind the Ivermectin from the wildtype and omicron variants are 265 pN and 198 pN for 10 A/ ns pulling speed respectively. This indicate, Ivermectin will be lesser effective towards omicron variant of the SARSCoV- 2 (Figure 11).
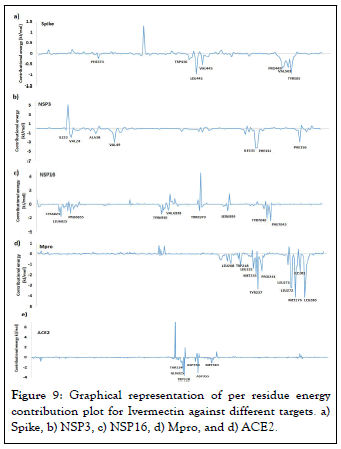
Figure 9: Graphical representation of per residue energy contribution plot for Ivermectin against different targets. a) Spike, b) NSP3, c) NSP16, d) Mpro, and d) ACE2.
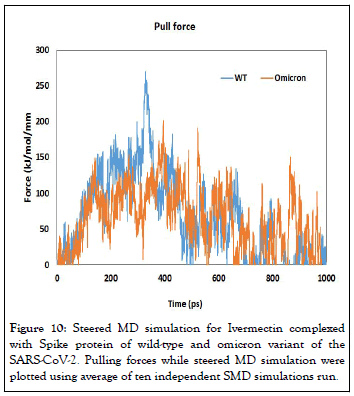
Figure 10: Steered MD simulation for Ivermectin complexed with Spike protein of wild-type and omicron variant of the SARS-CoV-2. Pulling forces while steered MD simulation were plotted using average of ten independent SMD simulations run.
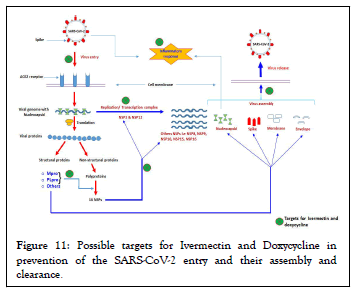
Figure 11: Possible targets for Ivermectin and Doxycycline in prevention of the SARS-CoV-2 entry and their assembly and clearance.
In search of effective therapeutics against COVID-19, different strategies have been applied and each therapeutic has different targets for the prevention of the disease severity [49]. Some drugs block virus spike interaction with host cell receptors or interfere with viral genome replication while some other drugs may obstruct the process of virus assembly. In the current study, drugs Ivermectin and Doxycycline were evaluated for their binding affinities against SARS-CoV-2 spike, Mpro, PLpro, RdRp, nucleocapsid, NSP3, NSP9, NSP10, NSP15, NSP16 and human ACE2 receptor by molecular docking. Further binding stability of spike, Mpro, NSP3, NSP16 and ACE2 protein complexed with Ivermectin were evaluated by MD simulation. In the last a comparison of wild-type and omicron variant of the SARS-CoV-2 was evaluated employing steered MD simulation by comparing their rapture force needed to unwind the Ivermectin with these variant spike protein.
During SARS-CoV-2 infection, interaction of spike-RBD protein with the host cell receptor facilitate virus invasion and determine viral tissue or host tropism [50]. Initial interactions of spike with host receptor (ACE2), and subsequent fusion of the host and viral membrane allows the viral genome to enter inside the host cells. It is now well established that, amino acid residues SER19, GLN24, THR27, PHE28, ASP30, LYS31, HIS34, GLU35, GLU37, ASP38, TYR41, GLN42, LEU45, LEU79, MET82, TYR83, ASN330, LYS353, GLY354, ASP355 and ARG357 of the human ACE2 interact with amino acid residues LYS417, GLY446, TYR449, TYR453, LEU455, PHE456, TYR473, ALA475, GLY476, GLU484, PHE486, ASN487, TYR489, PHE490, GLN493, GLY496, GLN498, THR500, ASN501, GLY502 and TYR505 of SARS-CoV-2 spike- RBD protein [51]. Our results show that, Ivermectin binds at the junction of spike-RBD and ACE2 interaction site indicating that it might have potential to inhibit the entry of the virus to the host cell (Figure 1A). Different amino acid residues of spike protein interacting with Ivermectin are ARG403, ASP405, ARG408, GLN409, GLY416, LYS417, TYR449, TYR453, LEU455, PHE456, SER494, TYR495, GLY496, GLN498 and TYR505 whereas amino acid interacting with ACE2 includes LYS26, LEU29, ASP30, ASN33 HIS34, GLU37, THR92, VAL93, GLN96, ALA386, ALS387, GLN388, PRO389, LEU392, ARG393, ARG559 and SER563 (Figure 1B). When molecular docking of Ivermectin was done SARS-CoV-2 spike protein of omicron variant it was observed that it has higher binding energy compared to the original wild-type variant indicting lower efficacy of the Ivermectin toward it (Figure 10). Further, it was confirmed by unbinding studying of Ivermectin with spike protein of wild-type and omicron employing SMD simulation. Both these finding predict that Ivermectin will be less effective for omicron variant at similar concentration of the Ivermectin.
It is well-known that ACE2 play vital role in regulating several physiological processes such as blood pressure, wound healing and inflammation. ACE2 is a multifunctional protein possessing carboxypeptidase, hydrolase, metalloprotease, and protease activity. So it is essential to known the effect of Ivermectin binding on the function of ACE2. A critical examination of the ACE2 shows that it active sites comprises of amino acid residues ARG169, ARG273, HIS374, 375, HIS378, GLU402, TRP477, LYS481, HIS505 and TYR515 for its various function [52,53]. Since Ivermectin does not block active site of ACE2 enzyme, it is unlikely to affect the normal physiological functions of this protein.
The RdRp is a fundamental component of SARS-CoV-2 replication/transcription machinery and it is one of the critical druggable targets for SARS-CoV-2. In our study it was observed that Ivermectin has significant binding energy (-9.1 kcal/mol) against the RdRp indicating that it may hamper the process of the viral genome replication and transcription. SARS-CoV-2 Mpro is considered as a promising druggable target because it plays vital role in viral gene expression and replication and are essential for maturation cleavage events of viral precursor polyprotein.
The NSP3 is the largest multi-domain protein encoded by the CoVs and is a vital component of the Replication/Transcription Complex (RTC). NSP3 function as an ADP-ribose-1’- phosphatase. Besides its macrodomain, NSP3 also contains a papain-like protease (PLpro) which is required during the processing of the CoVs NSPs. Both these SARS-CoV-2 proteins also showed significant binding with Ivermectin and doxycycline. Ubiquitination is a posttranslational modification that, among many other functions, is very important in the regulation of innate immune pathways [54]. PLpro is believed to antagonize the IFN response through its de-ubiquitinating enzyme activity, which allows the protease to remove ubiquitin from a substrate. Out findings show that, binding of Ivermectin with NSP3 and PLpro may block its enzymatic activity and thereby prevent SARS-CoV-2 from down-regulating the anti-viral interferon response. This may improve viral load clearance by host innate immune system and reduce the extent of viral pathogenesis without any previous memory of the antigen.
Viral RNAs are capped to safeguard their stability, efficient translation, and evading the innate immune system of the host cell. The RNA capping tactic are employed by several RNA viruses to avoid immune detection by Toll-Like Receptor 7 (TLR7) and Toll-Like Receptor 8 (TLR8) [55]. The viral RNA capping machinery is anatomically and mechanistically different compared to eukaryotic mRNA capping system. The CoVs NSP16 protein is a RNA cap modifying enzyme and become active when complexed with its activating partner NSP10 [56]. NSP16 binds with NSP10 resulting a complex which exhibits RNA cap (nucleoside-2′-O)-methyltransferase activity. This indicates that the binding of Ivermectin to the SAdenosylmethionine binding site will hamper the essential methyl-group transfer and inhibit the methyltransferase activity of NSP16. In the absence of methyl group attachment to the 5’cap of viral RNA, it will get easily detected by host innate immune system. Strong binding affinity of Ivermectin with NSP10 may also interfere in the formation of NSP16-NSP10 complex.
The CoVs N protein is a structural protein that forms complexes with viral genome, interacts with the viral membrane protein during virion assembly and plays an important role in enhancing the efficiency of virus transcription and assembly [57]. N-terminal region of nucleocapsid protein consists of positively charged amino acids, which are responsible for its RNA binding. CoVs NSP9 is most likely involved in viral RNA synthesis. NSP9 binds with RNA and interacts with NSP8 to execute its essential functions [58]. Dimeric form of NSP9 is important for viral infection. Any disturbance in the dimerization of NSP9 can be a way to overcome CoVs infection [59,60]. NSP15 is another viral proteins whose function is poorly defined but it was observed that a mutations in the active site of NSP15 abolish its endoribonuclease activity in vitro and reduces viral infectivity [61,62]. At the same time, any mutations outside of the active site are reported to have a larger effect on virus viability, indicating that NSP15 has role in CoVs infection apart from its function as an endoribonuclease [63,64]. Our molecular docking study shows that Ivermectin has superior binding with several druggable proteins compared to doxycycline. Altogether binding of Ivermectin with these proteins will hamper their individual function and decrease the viral genome replication, processing of virion packaging, and ultimately viral load.
Molecular docking study provides only information about the static poses of the most preferred conformations of a molecule in the binding pocket of a protein. Whereas, MD simulation is helpful in studying the stability of binding pose gained from docking, to restore the inclusive binding energy as well as energy contribution by different residues, to study the conformational variations happening in protein due to ligand binding/ unbinding etc. The MD simulation employed the classical Newtonian physics and give information about the actual movement and structural perturbations of a protein in its biological environment. Thus, MD simulation was performed to examine the stability and dynamics of protein to confirm the docking results and get more insight into the stability of ligandprotein complex. Our RMSD and RMSF study showed that the binding of the Ivermectin with different druggable targets of SARS-CoV-2 was stable and rigid. Intermolecular H-bonding between of Spike, Mpro, NSP3 and NSP16 with Ivermectin in their complex showed that the highest number of average Hbonds were formed between Spike and Ivermectin whereas least average number of H-bonds was observed in NSP3 and Ivermectin complex (Figure 7). Further, PCA analysis was performed using the MD trajectories which have been widely used to investigate and explore the global motion of a protein during the simulation. It enables us to study the influence of any varying parameters, by reducing the complexity of the collective motion which is associated with the phase space behaviour related to protein functions and stability. Our PCA study showed that all the studied protein complexed with Ivermectin have lower phase space compared to unbound proteins indicating more stability of the complex during the simulation. Compared to other targets complex, Spike and ACE2 have least phase space indicating higher stability of Ivermectin with viral spike and human ACE2 receptor proteins. Thus, our MD simulation study corroborated well with the findings of the molecular docking and it was observed that binding of Ivermectin with SARS-CoV-2 spike, Mpro, NSP3, NSP16 and human ACE2 receptor is very stable as observed my RMSD and RMSF study. Another important application of MD simulations is to calculate the thermodynamic of the proteinligand complex for determining the free binding energy. Since MM/PBSA is one of the most widely used and accepted methods for computing free binding energy, g_mmpbsa package was used to calculate it. The final binding energy obtained from MM/PBSA calculation was in well agreement with the findings of molecular docking study.
Most of the CoVs are known to bind with host metalloproteases (MMPs), predominantly for viral survival. Tetracyclines are known to chelate Zinc from MMPs [65]. Doxycycline being a tetracycline derivative, it will be beneficial in inhibition of SARSCoV- 2 infection because it chelates Zinc [66]. Tetracyclines also possess anti-viral activities and known to modulate innate immunity. Anti-viral activity of Doxycycline had already been reported against retroviruses and dengue virus [67,68]. They can decrease the expression of NF-kB and the release of inflammatory cytokines such as TNF-α, IL-1β and IL-6, independent of their antibiotic mechanism [69]. Thus, they could participate in limiting the ‘cytokine storm’ induced in COVID-19 patients. The lipophilic nature of tetracyclines and their strong pulmonary penetration could allow them to inhibit viral replication at cellular level.
Among Tetracyclines, Doxycycline has unique advantages such as long history of safety, the short duration of treatment and low cost. At the same time Ivermectin possess anti-viral activity for a range of the viruses. In their recent finding, Caly et al. found that Ivermectin significantly inhibited SARS-CoV-2 under in vitro conditions [33]. Very recent, clinical trials finding shows that combination of Ivermectin and Doxycycline has higher successful rate in COVID-19 patients compared to conventional therapy. Use of Ivermectin along with Doxycycline has fast recovery in patients who progress to more advanced stage of disease and reduced mortality rate in severe as well as critically ill patient [70]. Thus, the binding of the Ivermectin and Doxycycline with deferent druggable targets which help in preventing the SARS-CoV-2 entry and their assembly are depicted in the Figure 11.
In summary, our in silico mechanistic study shows that the possible mechanism behind the effects of Ivermectin and Doxycycline combination in COVID-19 disease may be due to inhibition of spike-ACE2 receptor interaction. Ivermectin will be lesser effective against current omicron variant of the SARSCoV- 2 for same dose. Apart from inhibiting viral entry, it might be helping in reducing the viral load by inhibiting RNA dependent RNA polymerase, ADP ribose phosphatase, endoribonuclease and NSP10-NSP16 complex mediated methyltransferase activities. Further in silico study required validation using wet lab experiments to make any conclusive mechanism underlying it.
Department of atomic energy, government of India.
The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Dharmendra Kumar Maurya, conceived, designed, performed and wrote the manuscript.
The author would like to thank Dr. S. Santosh Kumar and Dr. Deepak Sharma, radiation biology & health sciences division, Bhabha Atomic Research Centre, Mumbai for his valuable constructive suggestions and critical review of the article. The author also acknowledges the BARC supercomputer facility for MD simulation studies.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Maurya DK (2025) In Silico Exploration of Mechanisms Underlying the Synergistic Effects of Ivermectin and Doxycycline in the Treatment of SARS-CoV-2. J Clin Cell Immunol. 16:750.
Received: 23-Aug-2023, Manuscript No. JCCI-23-26235; Editor assigned: 25-Aug-2023, Pre QC No. JCCI-23-26235 (PQ); Reviewed: 08-Sep-2023, QC No. JCCI-23-26235; Revised: 11-Jul-2024, Manuscript No. JCCI-23-26235 (R); Published: 18-Jan-2025 , DOI: 10.35248/2155-9899.25.16.750
Copyright: © 2025 Maurya DK. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.