Advances in dairy Research
Open Access
ISSN: 2329-888X
+44 1300 500008
ISSN: 2329-888X
+44 1300 500008
Research Article - (2019)Volume 7, Issue 1
Bovine mastitis is a multifactorial disease that produces important losses in livestock and dairy industry. One of the main factors that causes this disease are microorganisms such as Staphylococcus aureus, which has multiresistance to antibiotics and virulence factors such as its ability to internalize into bovine mammary epithelial cells (bMECs), which allows bacteria to evade the host's immune system and persist inside the cell. However, coagulasenegative Staphylococci (CNS) have a significant role as bovine mastitis pathogens. S. agnetis is an emergent CNS pathogen isolated from bovine mastitis, whose capacity to internalize into bMECs is unknown. In the present work we characterized an isolate of S. agnetis from bovine mastitis, which was resistant to penicillin, dicloxacillin and ampicillin; also, the isolate showed resistance to methicillin through a mechanism that may be related to the presence of the mecA gene. In addition, S. agnetis internalized into bMECs (10% relative to S. aureus (ATCC 27543), which could be related to the presence of the fnbpA, clfB and spa genes. This is the first study that demonstrates the ability of internalization of S. agnetis into bMECs, which is a virulence factor that may represent a major problem in the management of bovine mastitis.
Mastitis bovine; Staphylococcus agnetis; Internalization
Mastitis is defined as the inflammatory response resulting of the infection and/or damage of the udder tissue, which is reported in numerous species, mainly in domestic dairy animals [1]. This pathology is the most frequent disease of dairy cattle and can be potentially fatal [2]. Mastitis is one of the diseases costliest to dairy industry associated with a reduced milk production, changes in milk composition and quality [3]. Staphylococcus aureus is a coagulasepositive bacterium, which is the main etiological pathogen of contagious bovine mastitis [4]. This microorganism is well known for its tolerance to wide range of antimicrobial agents and its ability to internalize and persist into bovine mammary epithelial cells, which allow it to evade the host immune system and survive inside a wide variety of mammalian cells that difficult the antimicrobial therapy [5]. Recently, coagulase-negative Staphylococci (CNS) have become the most common bovine mastitis isolates and are now predominant over S. aureus [6] and have been considered as emerging mastitis pathogens. In CNS group different microorganisms as S. sciuri, S. haemolyticus, S. chromogenes, S. epidermidis, S. saprophyticus, S. simulans and S. agnetis, among others, are included [7].
However, the information about the capacity of internalization of microorganisms of this group into bovine mammary epithelial cells is still limited. In particular, S. agnetis is a CNS bacterium isolated for the first time from bovine mastitis in 2012 [8] and later in 2015 in México [9]. Currently, the presence of S. agnetis in mastitis cases has been reported in various countries around the world including Brazil, USA, Canada and India [10-13].
However, to date there is no reported information on its ability to internalize into bovine mammary epithelium cells. The internalization of the bacteria in the host cell, is a virulence hallmark that makes difficult to treat the infection with conventional antibiotics, it also allows the bacteria to evade the host's immune system, as well as to persist inside the cell, which leads to the chronicity of infection and promotes its recurrence [14]. In order to increase the knowledge on this bacterium, in this work we characterized an isolate of S. agnetis obtained from subclinical bovine mastitis. Also, the internalization ability of this isolate into bovine mammary epithelium cells was evaluated.
Strain and microbiological culture
Staphylococcus aureus (ATCC 27543) previously isolated from a case of bovine clinical mastitis was used in this study as strain control. This strain has recognized capacity to internalize into bovine mammary epithelial cells [15]. S. agnetis 394696 isolate used in this work (NCBI/EMBL accession JQ 394696) was reported by León- Galván et al. [9]. Bacteria were grown at 37°C overnight in Luria- Bertani broth (DIBICO) for conducting experiments. The colony forming units (CFU) were adjusted by optical density determination (O.D. 600 nm).
Primary culture of bovine mammary epithelial cells (bMECs)
The isolation of bMECs was performed from alveolar tissue of lactating cow udders as described by Anaya-López et al. [16]. Cells from passages 4th to 7th were cultured in growth medium composed by DMEM medium/nutrient mixture F-12 Ham (DMEM/F-12K, Sigma) supplemented with 10% fetal calf serum (Gibco), 10 μg/mL insulin (Sigma), 5 μg/mL hydrocortisone (Sigma), 100 U/mL penicillin, streptomycin (100 μg/mL) and 1 μg/mL amphotericin B (Invitrogen). Subsequently, cells were grown in 5% CO2 atmosphere at 37°C.
Morphological and biochemical characterization of Staphylococcus agnetis 394696 isolate
In a previous work, S. agnetis 394696 was identified by colonial morphology, Gram staining and amplification of the 16S rDNA [9]. Additionally, in this work we carried out a biochemical characterization that included the coagulase test, mannitol fermentation and hemolysis production [17-19].
Antimicrobial susceptibility testing
The antimicrobial susceptibility test of S. agnetis 394696 was done using the Kirby-Bauer disk diffusion method in Muller-Hinton agar. The antimicrobial used were: Penicillin (PE) 6 μg, Dicloxacillin (DC) 30 μg, Pefloxacin (PEF) 5 μg, Cefuroxime (CXM) 30 μg, Gentamicin (GE) 120 μg, Cefotaxime (CTX) 30 μg, Sulfamethoxazole +Trimethoprim (SXT) 1.25 and 23.75 μg, Tetracycline (TE) 30 μg, Ampicillin (AM) 10 μg, Erythromycin (E) 15 μg, Ceftazidime (CAZ) 30 μg and Cephalothin (CF) 30 μg (BIORAD). Oxacillin resistance was determinate by agar dilution method in Muller-Hinton agar, supplemented with 4% NaCl and 6 μg/mL of Oxacillin (Sigma) after 48 h incubation a 37°C using ~ 5.4 × 106 CFU [20].
Internalization assays of Staphylococcus agnetis 394696 into bMECs
Internalization assays (gentamicin protection assay) were carried out as described by Ochoa-Zarzosa et al. [5]. bMEC monolayers (≈ 50,000 cells/well) were challenged with S. aureus or S. agnetis (MOI 30:1 bacterium per cell, to be able to compare with a bacterium with recognized internalization capacity). Briefly, bMECs were inoculated with bacterial suspensions and incubated for 2 h in 5% CO2 at 37°C.
After, bMEC monolayers were washed three times with PBS (pH 7.4) and incubated in grow media without serum, supplemented with 40 μg/mL gentamicin for 30 min at 37°C to eliminate extracellular bacteria. bMEC monolayers were detached with trypsin-EDTA (Sigma) and lysed with 200 μL of sterile distilled water. Then, bMEC lysates were diluted 100-fold, plated on LB agar for triplicate and incubated overnight at 37°C. The number of total CFU was determined by the standard colony counting technique. Data are presented as the percentage of internalization in relation to S. aureus strain.
Genomic DNA isolation
Bacteria were cultured in LB media for 16-18 h at 37°C; then, 1 mL of culture was mixed with lysis solution (SDS 10%, 0.6% Proteinase K) by vortexing and was incubated 1 h at room temperature. Genomic DNA was extracted by addition of 600 μL of phenol:chloroform: isoamyl alcohol (25:24:1); gently mixed for 2 min and centrifuged for 10 min at 16,000 g at 4°C. The aqueous phase was recovered, and the nucleic acids were precipitated with 600 μL of isopropyl alcohol. Finally, genomic DNA was diluted in 100 μL of sterile distilled water and stored at -20°C until use.
Amplification of fnbpA, clfB and spa genes of Staphylococcus agnetis 394696
Amplification of fnbpA, clfB and spa genes was carried out with a proof fidelity enzyme (Invitrogen). Oligonucleotides used and alignment temperatures are shown in Table 1. Amplification conditions were as follow: 3 min at 94°C; 30 cycles of 1 min at 95°C, 30 s at 59°C -62°C, 30 s at 72°C; and finally, 7 min at 72°C. An aliquot of 10 L of the PCR products was subjected to electrophoresis in 1.5% (w/v) agarose gels and stained with SYBR Green (Invitrogen) to amplicon visualization.
Biochemical and morphological characteristics of S. agnetis 394696 isolated from subclinical bovine mastitis
S. aureus is one of the most common etiological pathogens of contagious bovine mastitis, but in the last years, different bacteria have also become emerging pathogenic microorganisms of subclinical bovine mastitis including S. agnetis
In this sense, it was observed that S. agnetis 394696 isolate growth as circular colonies, convex, smooth, shiny and white that reach a diameter of 1-3 mm. Also, the S. agnetis 394696 isolate was negative for mannitol fermentation, non-hemolytic on bovine blood agar and coagulase-negative after 24 h (Figure 1).
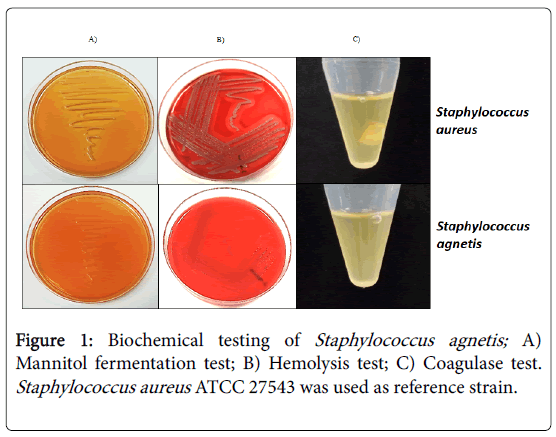
Figure 1: Biochemical testing of Staphylococcus agnetis; A) Mannitol fermentation test; B) Hemolysis test; C) Coagulase test. Staphylococcus aureus ATCC 27543 was used as reference strain.
The colony morphology of S. agnetis 394696 showed differences respect to the strain previously reported by Taponen [8], who reported that bacterium formed opaque and light gray colonies. This colony differences suggest that S. agnetis 394696 should has different characteristics and genetic variability that the strain reported previously [8].
This is not a strange phenomenon as several works have reported genetic variability and different properties between bacterium of the same genus and different species or same genus and species.
For example, has been reported that different isolates of S. aureus show variability in growth rate, colony size, and colony pigment; these differences have been associated with its virulence and antibiotic sensitivity [21].
On the other hand, the results of mannitol fermentation, production of hemolysis and coagulase test agree with a previous report [8].
Antimicrobial susceptibility patterns of Staphylococcus agnetis 394696
Due to the increase in the number of isolates of S. agnetis in subclinical bovine mastitis [8-13] the need to analyze its characteristics, virulence and pathogenicity is highlighted. To provide information about antibiotic susceptibility of S. agnetis 394696, an antibiogram was performed. S. agnetis 394696 was resistant to penicillin, dicloxacillin and ampicillin.
This result differs from data reported in India, where two collected isolates were resistant to only one antibiotic [13]. These profiles of resistance can be associated with the management practices and production systems present in the region.
This multi-resistance is an important factor that hinders the resolution of infection with conventional antibiotics. In this sense, infections caused by methicillin resistant staphylococci (MRS) are more harmful due to prolonged treatments and limited drug options because is intrinsically cross-resistant to virtually all of the β-lactam antibiotics [22].
In this sense, S. agnetis 394696 showed resistance to methicillin (oxacillin, 6 μg/mL) and harbor the mecA gene. Several studies suggested that mecA gene is present in all of the MRS [23]. However, some clinical isolates of S. agnetis [13] and other staphylococci like-S. aureus are mecA-positive but oxacillin-susceptible [24].
It has already been described that methicillin resistance is related to the expression of the protein penicillin-binding protein (PBP2), which is regulated by the mecA, mecR1-MecI system; however, if there is any failure in system activation, expression of the PBP2 protein decreases, thus, some isolates are methicillin-sensitive despite carrying mecA gene [23].
In S. agnetis 394696, the methicillin resistance can be attributed to classical mechanism of resistance related to this gene. Nevertheless, further studies are required to confirm the expression of PBP2 protein in S. agnetis 394696 (Figure 2).
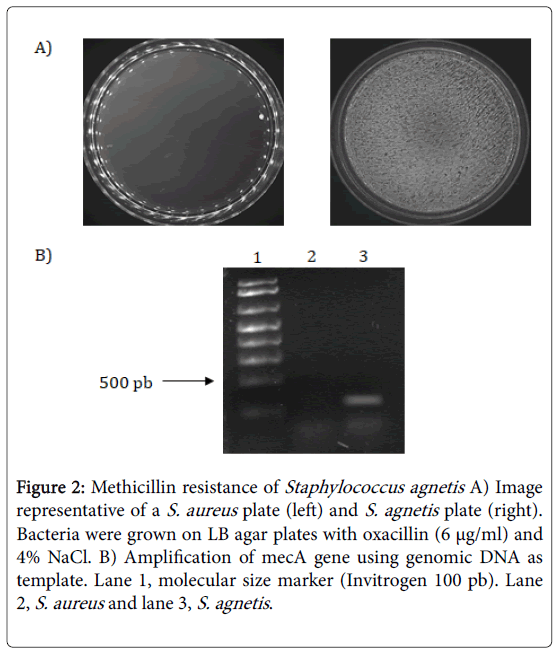
Figure 2: Methicillin resistance of Staphylococcus agnetis A) Image representative of a S. aureus plate (left) and S. agnetis plate (right). Bacteria were grown on LB agar plates with oxacillin (6 μg/ml) and 4% NaCl. B) Amplification of mecA gene using genomic DNA as template. Lane 1, molecular size marker (Invitrogen 100 pb). Lane 2, S. aureus and lane 3, S. agnetis.
Internalization of Staphylococcus agnetis 394696 into bovine mammary epithelial cells (bMECs)
S. aureus is one of the main causative agents of subclinical bovine mastitis due, among other factors, to the fact that some strains have the capacity to internalize into bovine mammary epithelium cells [15].
This phenomenon has already been described in Staphylococci coagulase-negative as S. epidermidis, which is associated with bovine intramammary infections [25].
However, previous to this work it was unknown whether S. agnetis has this characteristic. In the present work, internalization assays were carried out in a primary culture of bMECs and we used S. aureus ATCC 27543 as a control strain.
The results showed that S. agnetis 394696 internalized into bMEC 10% relative to control (Figure 3).
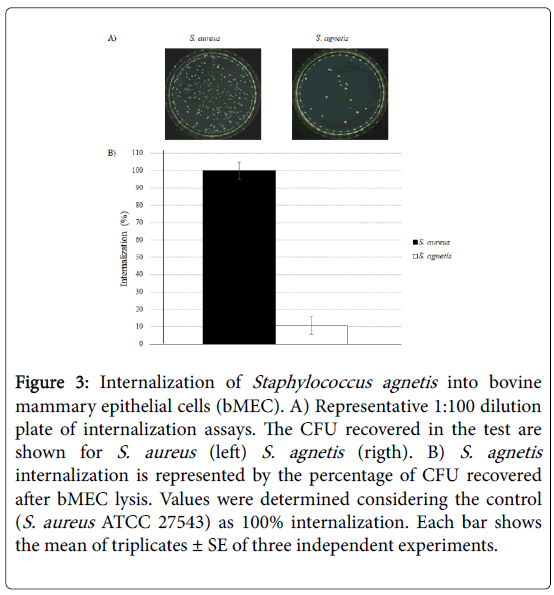
Figure 3: Internalization of Staphylococcus agnetis into bovine mammary epithelial cells (bMEC). A) Representative 1:100 dilution plate of internalization assays. The CFU recovered in the test are shown for S. aureus (left) S. agnetis (rigth). B) S. agnetis internalization is represented by the percentage of CFU recovered after bMEC lysis. Values were determined considering the control (S. aureus ATCC 27543) as 100% internalization. Each bar shows the mean of triplicates ± SE of three independent experiments.
With the above results, it is confirmed that, although with a lower percentage, S. agnetis has the ability to internalize into bMECs.
The most studied mechanism of internalization of S. aureus into nonprofessional phagocytic cells (c.a. bMECs) is through a process of receptor-mediated endocytosis (zipper mechanism) [14].
In this mechanism, the fibronectin binding protein (FnbpA) present on the bacterial surface binds to the fibronectin of the extracellular matrix, and this complex is recognized by the integrins of the host cell. On the other hand, clumping factor B (ClfB) is a bifunctional protein that binds to cytokeratin and fibrinogen, and protein A (SpA), is an abundant surface protein of S. aureus , which activates TNF receptors and EGF receptor (EGFR) signaling cascades that can perturb the cytoskeleton [26-29].
However, it is unknown if this mechanism is utilized by S. agnetis to internalize into bMECs. Using in silico analysis, it has been showed the presence of these genes in the genome of S. agnetis [30-32].
For this reason, we searched if S. agnetis 394696 harbor the fnbpA, clfB and spa genes. Interestingly, the expression of these genes was detected in S. agnetis isolate (Figure 4).

Figure 4: Expression of fnbA, clfB and spa genes in Staphylococcus agnetis. Analysis was performed using genomic DNA and PCR endpoint. fnbA gene (lane 2, S. aureus and lane 3 S. agnetis); clfB gene (lane 4, S. aureus and lane 5 S. agnetis); spa gene (lane 6, S. aureus and lane 7 S. agnetis). Lane 1, molecular size marker (Invitrogen 100 bp).
According to these results, S. agnetis has the ability to internalize into bovine mammary epithelium cells, which could be mediated by fnbpA, clf B and spa genes; however, further studies are necessary to demonstrate the role of these genes in this process.
Bovine mastitis remains as a problem of difficult treatment and resolution, mainly due to the variety of factors involved in the disease development (multidrug resistance and diversity of virulence factors of microorganisms). Here we showed that S. agnetis 394696 isolate is resistant to different antibiotics such as penicillin, dicloxacillin, ampicillin and methicillin. Also, to our knowledge this is the first report showing that this bacterium has the ability to internalize into bovine mammary epithelial cells. The resistant to different antibiotics and the capacity of S. agnetis to invade host cells are two crucial factors to consider in the management of bovine mastitis generated by this microorganism.
The authors thank students and colleagues of University of Guanajuato and Promoción de la Cultura y la Educación Superior del Bajío, A.C for their technical support during this study. The authors also appreciate the contribution of the dairy farmers to this study.
This research was supported by Grants from CONACyT 2017 (PEI-241362) and Consultoría en Biotecnología, Bioingeniería y Servicios Asociados, S.A de C.V. (COBBYSA).
Animal studies were carried out humanely and according with national and international Animal Care and Use Committee protocols.
All the authors declare no conflict of interest regarding this manuscript.
Citation: Gutiérrez Chávez AJ, Guzmán-RodrÃguez JJ, León-Galván FM, Barboza-Corona JE, Valencia Posadas M, Ochoa-Zarzosa A, et al. (2019) Staphylococcus agnetis: An Emergent Pathogen Isolated from Subclinical Mastitis with Capacity to Internalize into Bovine Mammary Epithelial Cells. J Adv Dairy Res 7: 221.
Received: 11-Mar-2019 Accepted: 19-Mar-2019 Published: 27-Mar-2019 , DOI: 10.35248/2329-888X.19.7.221
Copyright: © 2019 Gutierrez-Chavez AJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.