Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2009) Volume 2, Issue 11
Gonococcal infection and transmission is a global health problem and till date no effective vaccine is available to prevent the disease transmission. Recently, analysing membrane proteome of N. gonorrhoeae we have shown that, several membrane associated proteins of the gonococcus might be important in developing anti-gonorrhoea drugs. Here, we explored the possibility of such 19 essential membrane transporter targets those may be useful in developing peptide vaccines too. Using a classical in silico technique, we have identified four best epitopes from three transporters. All identified epitopes are antigenic and predicted to induce both the T- and B-cell mediated immune responses and transmembrane helix prediction shows that, selected epitopes are mostly outside of the membrane that indicates the suitability of these epitopes to be potential peptide vaccine candidates. Identified epitopes require experimental validation.
Keywords: N. gonorrhoeae, Peptide vaccine, Essential membrane transporters, Drug targets.
Gonorrhoea caused by Neisseria gonorrhoeae is the second most common sexually transmitted disease (STD) in USA and the commonest STD prevalent in underdeveloped countries (Centers for Disease Control and Prevention, 2007; Cornelissen, 2008). The disease is mostly found in women where polyarthralgia, tenosynovitis, arthritis are common symptoms (Bardin, 2003). Severe infection occasionally causes pelvic inflammatory disease (PID) (Furuya and Tanaka, 2009) that leads to infertility and ectopic pregnancy due to permanent blockage of the fallopian tube (Tapsall, 2006). In men, common symptoms are urethritis, epididymitis, and prostatitis (Furuya and Tanaka, 2009). The mode of disease transmission is generally from infected man to a woman and till date this gonococcal transmission and infection is a global public health problem due to lack of appropriate vaccine against the pathogen and the immerging antibiotic resistance of the gonococcus (Snyder et al., 2001; Workowski et al., 2008).
Although, previous researches have identified a number of candidate vaccines from membrane and cell wall associated proteins such as pili proteins (Rothbard et al., 1985), opa, lipooligosaccharides, protein-I, lactoferrin (Lbpl, Lbp2), IgA1 proteases (Barbosa-Cesnik et al., 1997), 2C7 oligosaccharide (OS) epitope (Gulati et al., 2001), protein-IB (PorB) (Zhu et al., 2004), phospholipase A (PldA) (Bos et al., 2005), and transferrin-binding proteins (TbpA and TbpB) (Price et al., 2005; Thomas et al., 2006; Price et al., 2007); none of these found effective in practice. Therefore, there is a need of new vaccine candidate identification and validation.
In our recent study, using subtractive genome analysis, we found that several membrane associated essential transporters might be good drug targets against the pathogen (Barh and Misra, 2009). As these targets are associated with cell membrane and few of them are also exposed to cell surface/cell wall, these targets may also be suitable for vaccine designing. Hence, in this study, we made an effort to identify such essential gonococcal transporters those can be used as drug targets as-well-as designing of peptide vaccines.
Selection of Essential Transporters
From our previous report (Barh and Misra, 2009), membrane and cell wall associated 19 essential transporters (putative drug targets) of N. gonorrhoeae were selected for this study. Important transporters comprise of ABC transporter permease proteins, lipoprotein carriers, antibiotic resistance efflux pump components, transferrin-binding protein A, preprotein translocase subunits, and F0F1 ATP synthase subunits. Assuming that the virulent transporters will be more antigenic and will produce better immune response, we used Virulence Factors of Pathogenic Bacteria Database (VFDB) (Chen et al., 2005) to identify such transporters form our selected pool of essential 19 transporters.
Epitope Identification
To identify epitopes, a classical strategy was taken where the identified epitopes should be antigenic and have the ability to induce both the T-cell and B-cell mediated immunity. Briefly, amino acid sequence of each transporters was retrived from Swiss-Prot protein database (http://us.expasy.org/sprot) and subsequently analyzed for antigenicity using B-cell antigenic site prediction server “Antigenic” (http://bio.dfci.harvard.edu/Tools/antigenic.pl). Using the default parameters of both the BCPred and AAP prediction modules of BCPreds (ELManzalawy et al., 2008), B-cell non-overlapping epitopes were also identified from each transporter. Primary selection of the predicted B-cell epitopes were done based on the scores where the cut off values for "Antigenic" and BCPreds were respectively 1.1 and 1.0. In the next step, overlapping epitope sequences from “Antigenic” and BCPreds were selected and sequences were aligned to get a continuous stretch of amino acid sequence that possess both antigenic sequences as-well-as the B-cell binding sites. This continuous stretch of the antigenic B-cell epitope was then analyzed using ProPred-1 (Singh and Raghava, 2003) and ProPred (Singh and Raghava, 2001) with default parameters to identify respectively MHC class I and MHC class II binding epitopes. Selected numbers of MHC binding alleles were 47 and 51 respectively for MHC class I and II. Proteosomal cleavage sites of identified epitopes were also analyzed. Epitopes that can bind both the MHC classes and maximum MHC alleles were selected. The final selection of epitope sequences were done based on the criteria that the epitope sequence should have antigenic B-cell epitope binding sequences as-well-as both the MHC classes binding sequences. Therefore, the selected sequences, in this way, have the ability to generate both the B-cell and T-cell mediated immune responses. The strategy of the epitope prediction is represented in Figure 1. The final list of epitopes was prepared based on the position of the epitopes in respect to the signal peptide and transmembrane (TM) helices using the TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Krogh et al., 2001) and Phobius (http://phobius.sbc.su.se/) (Käll et al., 2004) transmembrane topology prediction servers. Epitopes those are exposed to cell surface/out side of the membrane were selected.
Virulent Transporters
Only two virulent essential transporters namely ABC transporter iron-uptake permease inner membrane protein (FbpB) [GenBank: NGO0216] and Transferrin-binding protein A (TbpA) [GenBank: NGO1495] were identified from these 19 essential transporters selected in this study. While, FbpB is an integral membrane protein; TbpA is found to be a cell outer membrane protein. Both the transporters are involved in inorganic ion transport (Table-1 marked with an asterisk *).
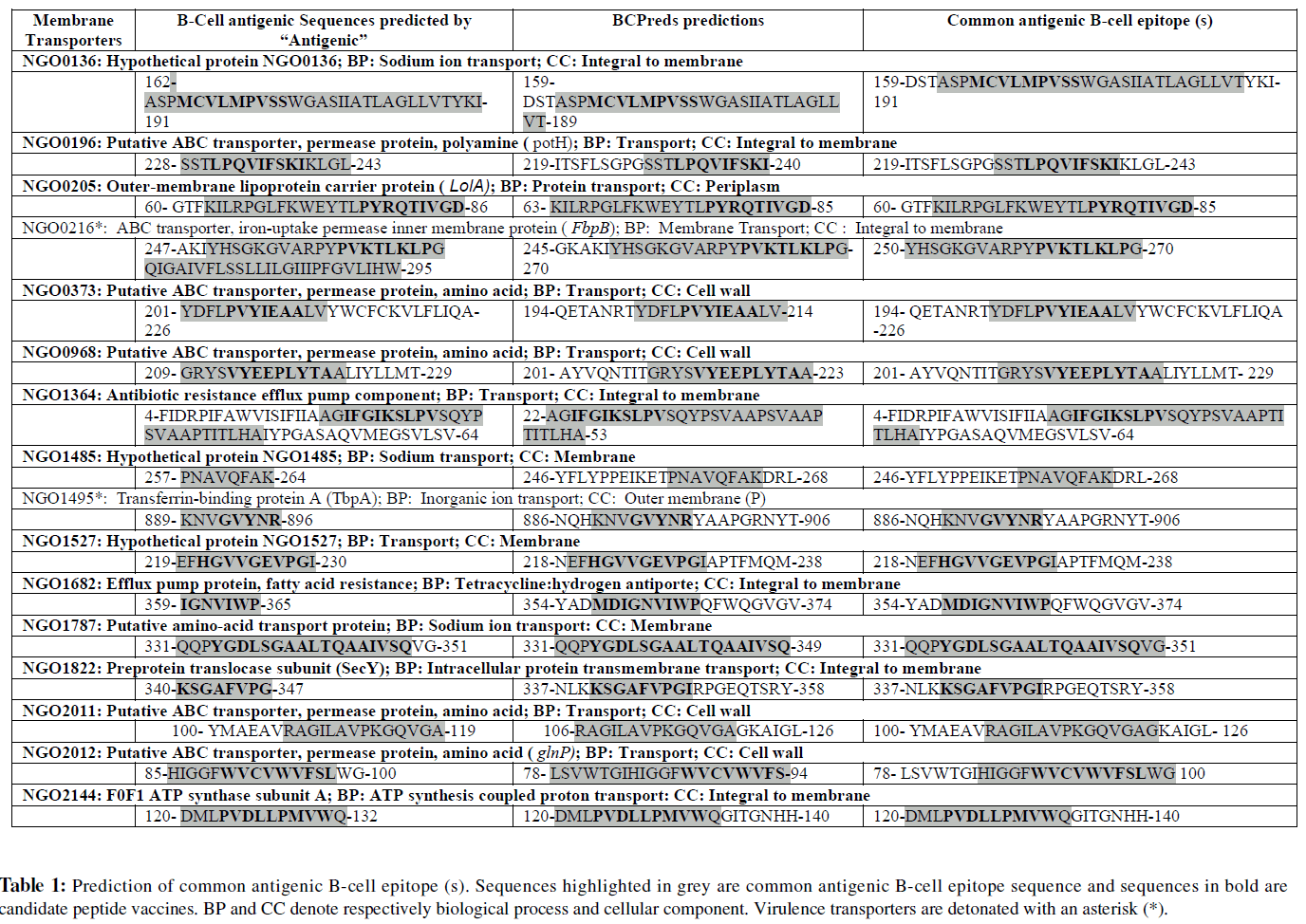
Identified Antigenic B-cell Epitopes
Following the method as mentioned earlier, common antigenic B-cell epitopes for each transporter were identified using “Antigenic” and BCPreds. It was found that, the predicted Bcell epitope sequences by BCPred and AAP prediction modules of BCPreds highly varied and therefore the common sequences generated by these two algorithms of BCPreds were considered in most cases to select B-cell epitopes. The variable lengths of antigenic sequences generated by “Antigenic” and the 20 mers length B-cell epitope sequences predicted by BCPreds were then analysed to find common B-cell antigenic epitope sequences those are listed in Table 1. Three transporters LolC [GenBank: NGO0769], Sec A [GenBank: NGO0996], and ATP synthase subunit B [GenBank: NGO2146] did not show any overlapping common sequences of antigenic B-cell epitope. Therefore, these three transporters were excluded form further analysis.
Identified Candidate Peptide Vaccines
Selected antigenic B-cell epitope sequences were analysed with ProPred 1 and ProPred to identify respectively MHC I and MHC II binding T-cell epitopes. The common T-cell epitope sequences those bind to both the MHC molecules were selected. It was found that Hypothetical protein which is involved in sodium transport [GenBank: NGO1485] and ABC transporter permease protein [GenBank: NGO2011] do not have any common T-cell epitope sequences for their respective antigenic B-cell epitope sequences predicted in this study. Therefore these two transporters were found not suitable for vaccine development. The final outcome of this study was 15 epitope sequences from 14 transporters. Predicted peptides those may be considered for vaccine development based on MHC allele binding ability are listed in Table 2. It has been found that epitopes from ABC transporter permease protein [GenBank: NGO2012] and Hypothetical protein [GenBank: NGO0136] have the highest numbers of MHC allele binding ability (71 and 60 respectively). But the epitopes from the virulent transporters FbpB [GenBank: NGO0216] and TbpA [GenBank: NGO1495] showed moderate number of MHC allele binding probability (26 and 39 respectively). The lowest number of MHC allele binding ability was found by the epitope sequence from the Hypothetical protein [GenBank: NGO1527] which is only 15.
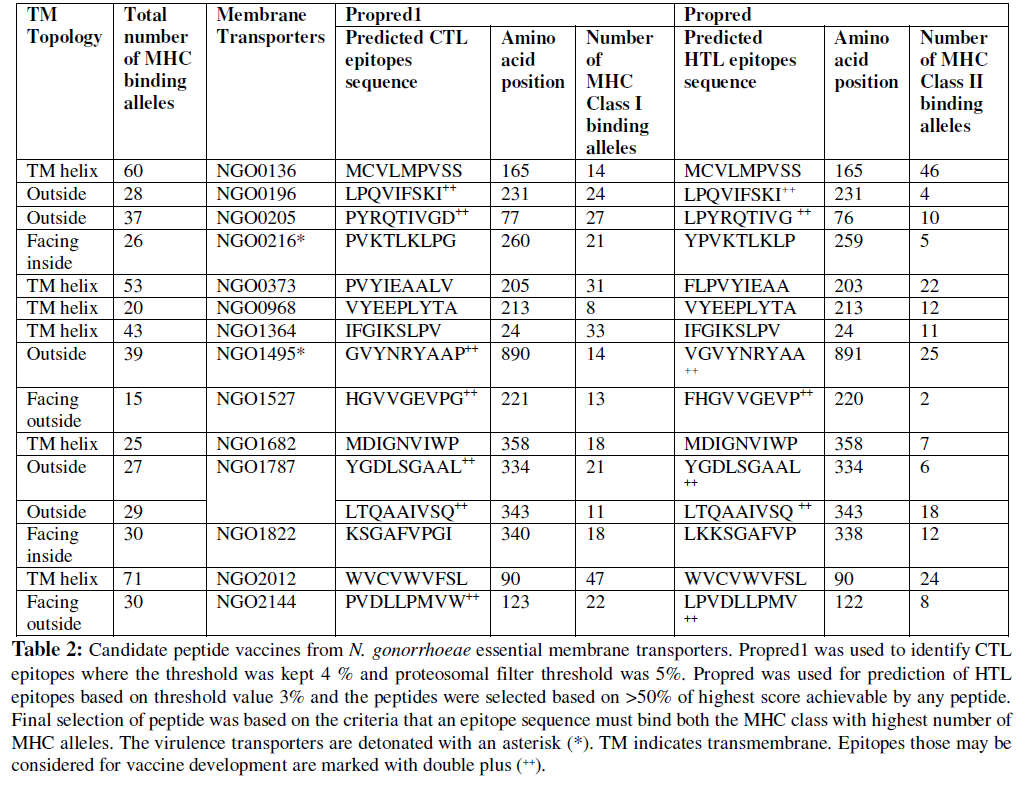
A peptide that can be considered for peptide vaccine must be surface localized or exposed to the cell outer environment. Therefore, these listed epitopes in Table-2 were further screened to determine whether an epitope is a transmembrane (TM) helics or exposed to the cell outer surface using TMHMM 2.0 (Krogh et al., 2001) and Phobius (Käll et al., 2004). The analysis revealed that, selected epitopes from Hypothetical protein [GenBank: NGO0136], FbpB [GenBank: NGO0216], ABC transporters [GenBank: NGO0373 and GenBank: NGO0968], Antibiotic resistance efflux pump component [GenBank: NGO1364], fatty acid resistance efflux pump protein [GenBank: NGO1682], SecY [GenBank: NGO1822], and GlnP [GenBank: NGO2012] are either TM helices or facing inside to the cell. Hence selected epitopes from these transporters are not suitable in developing peptide vaccine. Therefore, the final outcome of this study i.e. the vaccine candidates are seven epitopes from six transporters viz. ABC transporter (PotH) [GenBank: NGO0196], LolA [GenBank: NGO0205], TbpA [GenBank: NGO1495], Hypothetical protein [GenBank: NGO1527], Amino-acid transport protein [GenBank: NGO1787], and ATP synthase subunit A [GenBank: NGO2144] (Table-2, marked with double plus ++).
Attempts to develop an effective anti-gonorrhoea vaccine are not yet fully successful. Therefore, in search of candidate new peptide vaccines, we have analyzed 19 membrane associated essential transporter drug targets of N. gonorrhoeae to elucidate the possibility of these transporters to be used for dual purpose (as drug target and vaccine candidate). The current analysis shows that six essential membrane transports may be used for this purpose. To be a good peptide vaccine, a peptide should be able to activate both the helper T-lymphocytes /CD4+/ MHC II and CD8+ cytotoxic T- lymphocytes/ MHC-I (Pancré et al., 1996; Singh and Raghava, 2001). Therefore, in this study we made an effort to identify such peptides those are antigenic and are also able to maximum number of both the MHC class I and class II alleles. Fifteen peptides have been ident ified from 14 transports those can bind to maximum number of MHC alleles and have the potentiality to induce both the B-cell and T-cell mediated immunity. But 8 epitopes are found either TM helixes or facing towards cell inside therefore these epitopes are not suitable for vaccine designing. Hence, the rest seven epitopes form six transporters are selected as possible candidate peptide vaccines.
ABC transporters have previously been reported as good targets for vaccine development against pathogenic bacteria (Garmory et al., 2004). In this study we found that, epitope “LPQVIFSKI” from ABC transporter (PotH) [GenBank: NGO0196] is exposed to the surface and can bind to total 28 MHC alleles. Similarly, epitope “PYRQTIVGD” from Outermembrane lipoprotein carrier protein (LolA) [GenBank: NGO0205] found to interact with 37 MHC alleles. Therefore, these two peptides may be considered for vaccine development against the gonococcus.
Essential sodium ion transporters in bacteria have been proposed suitable for both drug and vaccine targets (Häse et al., 2001). Two epitopes viz. “YGDLSGAAL” and “LTQAAIVSQ” from Amino-acid transport protein [GenBank: NGO1787] respectively bind to 27 and 29 MHC alleles and are exposed to outside of the membrane (Table 2). Therefore, these two epitopes may be tested to develop anti- gonorrhoea vaccine.
Peptides from iron uptake ABC transporters are found to immunize mice against Streptococcus pneumoniae (Brown et al., 2001) and gonococcal iron transport systems have been reported to be potential vaccine antigens (Cornelissen, 2008). The transferrin- binding protein A (TbpA) [GenBank: NGO1495] in this study is a virulent protein and also a candidate drug and vaccine target (Price et al., 2005; Barh and Misra, 2009). This analysis shows that epitopes selected from TbpA have a partial antigenic sequence “GVYNR”; although the epitopes from this transporter “GVYNRYAAP” and “VGVYNRYAA” having the antigenic sequence are able to bind total 39 MHC alleles. Therefore, vaccines developed based on these peptide sequences may not be able to produce optimum immune response.
Other potential selected epitopes “HGVVGEVP” and “PVDLLPMVW” respectively from Hypothetical protein NGO1527 [GenBank: NGO1527] and ATP synthase subunit-A [GenBank: NGO2144] may not be suitable for developing vaccine as they are partly TM helices facing towards outside, although they are found to bind respectively 15 and 13 MHC alleles.
In this study, we have identified four most probable candidate peptide vaccines from three membrane transporters drug targets of N. gonorrhoeae out of 19 targets tested. These three transporters therefore may be useful in developing drug as-well-as vaccines against the pathogen. Predicted epitopes are antigenic and have potentiality to induce both the T-cell and B-cell mediated immunity. These candidates may be superior to those previously identified vaccine candidates. Experimental validation of these epitopes is required.
Financial Support
This research was carried out without any financial support or grant.
We are thankful to all IIOAB members for their moral support and encouragement while carrying out the analysis.