Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2024)Volume 17, Issue 1
Computation study of target genes has proven to be useful in contemporary research. This research was carried out to assess variation in rbcl gene of African yam bean (Spenostylis stenocarpa) and related legumes in the Fabaceae family. Sequences of rbcL gene were downloaded from National Center for Biotechnology Information (NCBI) database and used for this study. Multiple sequence alignment was carried out on the nucleotide sequences and all gaps were excluded from the analysis. The results of the phylogenetic analysis grouped the legumes into two major clades, while the maternal limeage analysis showed that African yam bean must have originated from Cajanus cajan (pigeon pea) and Glycine max (soy bean). The Guauine-cytosine content of the plants was similar and the domain detected in the sequences was low complexity which was the same in all the legumes into two major clades, while the maternal lineage analysis showed that African yam bean must have originated from Cajanus cajan (pigeon pea) and Glycine max (soy bean). The Guanine Cytosine (GC) content of the plants was similar and the domain detected in the sequences was low complexity which was the same in all the legumes sampled in this study. The present study suggests high similarly among the legumes based on information on their rbcl gene.
rbcL gene; Bioinformatics; African yam bean; Spenostylis stenocarpa
African Yam Bean (AYB) is a leguminous crop of tropical African origin belonging to the family Fabaceae, which is the second biggest and one of the most economically important families among the dicotyledons [1]. African yam bean was believed to originated in Ethiopia and both wild and cultivated species are found in tropical Africa as far south as Zimbabwe in East Africa from Northern Ethiopia (Eritrea) to Mozambique including Tanzania and Zanzibar [1]. It is also cultivated throughout West African countries particularly Nigeria Cameroun, Ghana, Togo and Cote d'Ivoire. African yam bean tolerates a wide range of climatic, geographical and edaphic ecologies [2]. It is capable of growth in marginal areas where other pulses fail to thrive. It survives well in weathered soils where rainfall could be extremely high, tolerates acidic, leached and infertile soil. The crop is an annual grain legume and has a pattern of growth similar to those of other grain legumes [3].
The center of diversity affirms that African yam bean spreads from the West through the East and Southern parts of Africa according to Germplasm Resources Information Network (GRIN, 2009) [4]. These areas host the genetic resources of African yam bean. The utilization of African yam bean has links with the socio-cultural values of some ethnic groups within the African society. For instance, the Avatimes in Ghana prepare a special meal from it during the celebration of puberty rites of adolescent girls. Likewise, a special meal from it features during the marriage ceremony among the Ekiti people in Nigeria, pointing that the different forms of local recipes are prepared from the crop to meet the dietary needs of the people [5]. In many areas where AYB it is cultivated, it is regarded as a security crop for fallow farmlands in the preparation for a new planting season and primarily consumed as staple crops.
Despite the economic importance of AYB, there is a decrease in agro-biodiversity of this species in many parts of Nigeria probably due to lack of awareness of the potential of this neglected crop, poor methods of propagation, processing, marketing and consumption of the crop. This crop has also undergone little or no genetic improvement to boost its agronomic and nutritional qualities. Though rich in nutrients, it is still largely underexploited compared to other legumes such as cowpea (Vigna unguiculate), pigeon pea (Cajanus cajan), common beans (Phaseolus vulgaris), soybeans (Glycine max) and Adzuki beans (Vigna angularis) [6-8].
An understanding of the genetic make-up in crops as well as its evolutionary relationship and diversity to its related legumes may result to genetic improvement and utilization [9,10]. This can be achieved by the application and utilization of molecular/ sequence data. Presently, many chloroplast, mitochondrial and nuclear genes have been utilized for studying and understanding sequence variations and evolutionary trends at the genus level [11,12]. One of such chloroplast gene is Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (RuBisCO) Large, rbcL gene has been reported to be useful for the study of phylogenetic relationship of flowering plants at the species and generic level [13-16]. The RuBisCO large subunit (rbcL) plastid marker was commonly used to study unknown taxonomic position of species to elucidate taxonomic connections between different species [17].
The Ribulose Biphosphate Carboxylase (rbcL) sequence method has been extensively used in studies of evolution, phylogeny, biogeography, population genetics and systematics because it can be readily copied and not strikingly different for related species [18-20]. The sequence of rbcL has been recorded in many studies and it is clear that this marker has great potential and benefit in terms of studying the genetic variations of the natural populations [21,22]. It has also been demonstrated that rbcL primers are useful for inter- and intraspecific evolutionary studies of plants [14].
The expression in silico was first used in public in 1989 in the workshop “Cellular Automata”: Theory and Application in Los Alamos New Mexico by Pedro Miramontes a mathematician from national autonomous University of Mexico (UNAM) who presented the report “DNA and RNA physiochemical constraints, Cellular Automata and Molecular Evolution’’. Plants genome shows a wide array of architectures i.e. (genetic make-up) varying immensely in size, structures and content. Some organelle DNAs have even developed elaborate peculiarities such as scrambled coding regions, non-standard genetic codes and convoluted modes of post-transcriptional modification and editing all of which has been deciphered using bioinformatics tools.
Bioinformatics is an interdisciplinary field that develops methods and software tools for understanding biological data. As an interdisciplinary field of science, bioinformatics combines computer science, biology, mathematics and engineering to analyze and interpret biological data. Bioinformatics has been used for in silico analyses of biological data and genes using mathematical and statistical techniques. Bioinformatics has become an important part of many areas of biology. In experimental molecular biology, bioinformatics techniques such as image and signal processing allow extraction of useful results from large amount of raw data. In the field of genetics and genomics, it aids in sequencing and annotating genomes and their observed mutations. It plays a role in the text miming of biological literature and the development of biological and gene ontologies to organize and query biological data. It also plays a role in the analysis of gene and protein expression and regulation. Bioinformatics tools aid in the comparison of genetic and genomic data and more generally in the understanding of evolutionary aspects of molecular biology. At a more integrative level, it helps to analyze and catalogue the biological pathways and networks that are an important part of systems biology. In structural biology, it aids in the simulation and modeling of Deoxyribo-Nucleic Acid (DNA), Ribo-Nucleic Acid (RNA), proteins as well as bimolecular interactions. In Genetics and Biochemistry, in silico studies can be used to examine the molecular modeling of gene, gene expression, gene sequence analysis and 3D structure of proteins, identification of diseases and prediction of lipid metabolic pathways.
This research is therefore designed to use bioinformatics tools to characterize the rbcL gene in AYB and some selected legumes.
Despite the numerous benefits of African yam bean in agricultural productivity and food security, it suffers several limitations when compared to other legumes ranging from the presence of high anti-nutritional factors (tannins, saponins, oxelate, phytate, trypsin inhibitor and lectin), hardness of seed- coat and hence longer time to cook, presence of secondary metabolites, lengthy lifecycle, photoperiodic sensitivity and low yields [23-26]. Up until now, the genetic diversity present in AYB remains poorly understood. Chloroplast gene markers are considered useful tools to trace demographic history, explore species divergence and identify species. One of such Chloroplast gene markers universally implored in genetic diversity studies is Ribulose Biosphosphate Carboxylase Large (rbcL) gene given its universality, ease of amplification and alignment. This research was designed to provide information on the genetic diversity by characterizing rbcL gene in AYB and some selected legumes. The aim of this study is to characterize the rbcL gene in AYB and some selected legumes.
Experimental site
The in silico study was carried out in the bioinformatics laboratory of the department of genetics and biotechnology, University of Calabar, Calaabar.
Retrieval of nucleotide and amino acid sequences
Nucleotide sequences of African yam bean and five related legumes in the family of fabaceae were downloaded from gene bank at National Centre for Biotechnology Information. This was done by downloading the FASTA format for the nucleotide sequences. The Genbank accession numbers of the nucleotide sequences were; Sphenostylis stenocarpa (OK254196.1, OK254195.1, OK254194.1, OK254193.1 and OK254192.1), V. unguiculata (OK104832.1, OK104823.1, OK104822.1, OK104821.1 and OK104820.1), Vigna angularis (MH391977.1, MH391976.1, MH391975.1, MH391974.1 and MH391973.1), Cajanus cajan (AB045791.1, AB045790.1, Z95535.1, OP710494.1 and MH549758.1), Phaseolus vulgaris (MN078233.1, MN078232.1, MN078231.1, MN078230.1 and MN078173.1) and Glycine max (Z95552.1, LC743621.1, OK104831.1, OK104830.1 and MW929340.1).
Phylogenetics and maternal lineage
The phylogenetic relationship between African yam bean and the selected related legumes was determined using Molecular Evolutionary Genetic Analysis (MEGA 6) software. Nucleotide sequences download from Genbank were aligned using multiple sequence alignment of ClustalW excluding all the gaps. The phylogenetic grouping was based on 1000 bootstrap replicates. The maternal lineage was also traced using MEGA 6.
Estimation of guanine-cytosine content of the rbcl gene
The guanine-cytosine content of the rbcl gene was predicted using a Genscan online software where the Fast Adaptive Shrinkage Threshold Algorithm (FASTA) format of the nucleotide sequences were pasted in the space provided and the genscan ran. The percentage of GC content was recorded for each legume sample.
Domain architecture
The domain architecture was determined using Specific, Measurable, Achievable, Relevant and Time-bound (SMART) online software. Tis was performed using the FASTA format of the amino acid sequences of the rbcl downloaded from Genbank. The different domains, the start and end regions were recorded for each legume used in this study.
Phylogenetics and maternal lineage
The phylogenetic relationship between African yam bean (S. stenocarpa) and the related legumes used in this study is presented (Figure 1). The result revealed that the legumes were divided into two clades based on the phylogemy. The first major clade comprised of two legumes (G. max and C. cajan). The second major clade had two sub-clades. Each sub-clade within the second major clade had samples of the same species of legume grouped together except S. stenocarpa with accession number: OK254194.1 which was grouped under a different sub-clade. Generally, the phylogenetic free revealed that individuals of the same species of legume were grouped together with specific sub-clades without intermixing or introgression. The results of the maternal lineage as presented, shows that the origin of S. stenocarpa (African yam bean) could be traced to C. cajan and G. max (Figure 2).
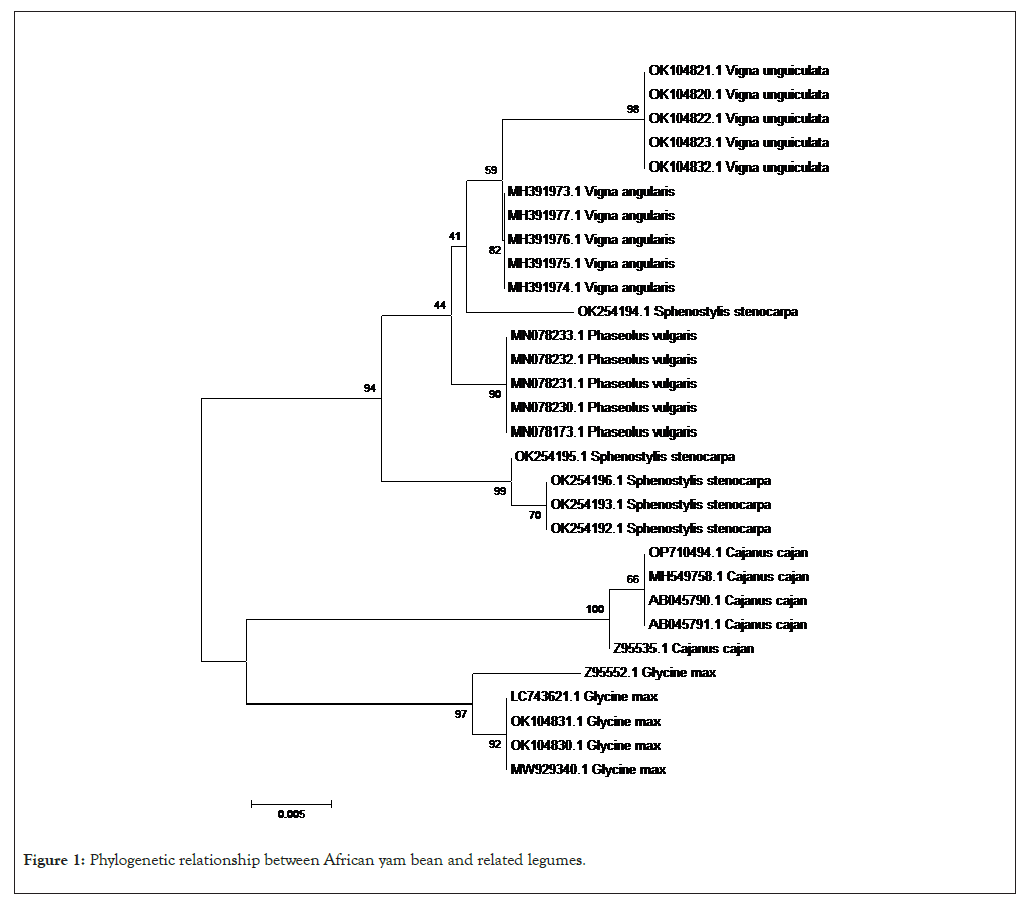
Figure 1: Phylogenetic relationship between African yam bean and related legumes.
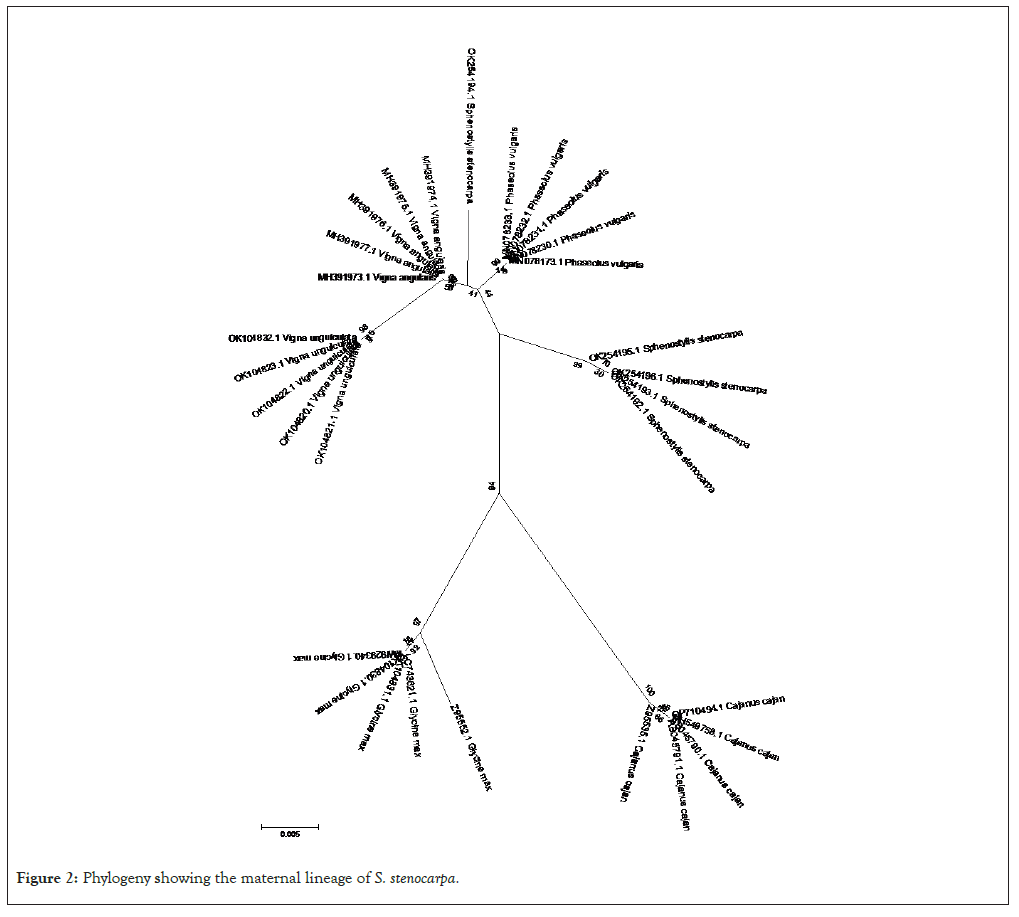
Figure 2: Phylogeny showing the maternal lineage of S. stenocarpa.
Guanine – cytosine content
The percentage of guanine-cytosine content on the nucleotide sequence of rbcl gene is presented (Figure 3). The highest GC content was observed from rbcl of G. max (44.69%) followed by V. angularis and P. vulgaris which both had the same GC content of 43.58%. The lowest G-C contents was recorded in rbcl gene of C. cajan (43.14%) (Figure 3).
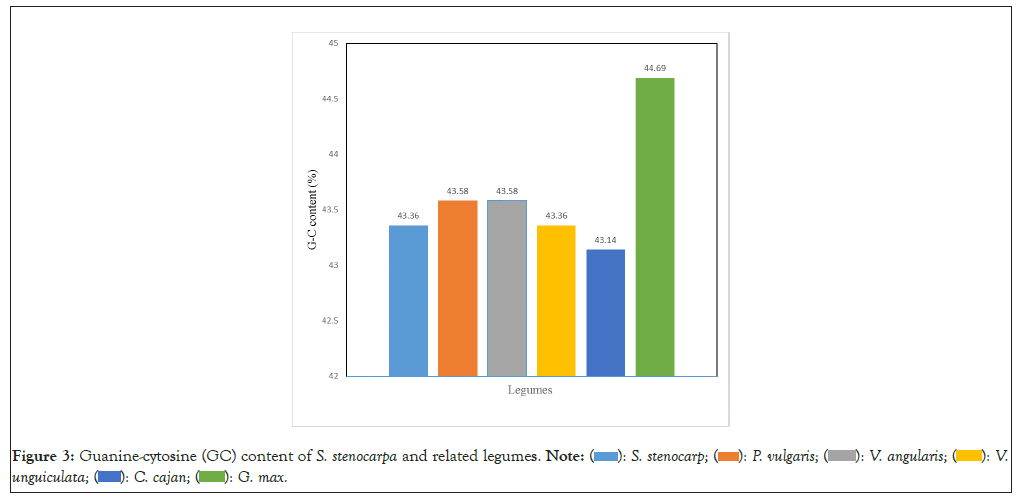
Figure 3: Guanine-cytosine (GC) content of S. stenocarpa and related legumes. 
Domain architecture
The different domains detected in the rbcL protein sequences of the legumes used in this study are presented. It was observed that all the legumes had the same type of domain as low complexity. All the legumes had two low complexity domains each except C. cajan and G. max which both had a single low complexity domain (Table 1).
| Species | Domains | Start | Stop |
|---|---|---|---|
| S. stenocarpa | Low complexity | 8 | 29 |
| Low complexity | 67 | 78 | |
| P. vulgaris | Low complexity | 9 | 28 |
| Low complexity | 67 | 78 | |
| V. angularis | Low complexity | 8 | 30 |
| Low complexity | 68 | 79 | |
| V. unguiculata | Low complexity | 8 | 30 |
| Low complexity | 68 | 79 | |
| C. cajan | Low complexity | 13 | 30 |
| - | - | - | |
| G. max | Low complexity | 9 | 29 |
| - | - | - |
Table 1: Domain architecture in rbcl protein sequences of African yam bean and related legumes.
Phylogeny is the progression or historical development of a plant, animal or microbial species and the tracing of such similar groups having a common ancestor. It is a visual graphical representation of the evolutionary relationship between different organisms, showing the path through evolutionary time from a common ancestor to different descendants [27]. Similarities and divergence among related biological sequences revealed by sequence alignment often have to be rationalized and visualized in the context of phylogenetic free. But with the development and use of computational and an array of bioinformatic tools, the ability to analyze large data sets in practical computing times and yielding an optimal or near optimal solutions with high probability are being possible [27]. In this study bioinformatic tools were used to evaluate the phylogenetic relationship between African yam bean (S. stenocarpa) and related legumes from fabaceae family. The phylogenetic free divided all the legumes into two major cluster, however, African yam bean was sub-grouped with Phaseolus vulgaris, Vigna argularis and Vigna unguiculata. This suggest that the evolutionary charges that have occurred in the sequences of the legumes are similar. The results of the phylogenetic relationship of the legumes which generated two major clusters is similar to the report of who also reported two major clusters in the phylogenetic relationship of legumes in fabaceae plants based on DNA barcoding with subsequent division into four subgroups [28]. In corroboration to the present findings also reported two major clades in a study on phylogenetic analysis of Genista l. (Fabaceae species) from Turkey based on inter-simple sequence repeat amplification [29].
The results of the maternal lineage analysis in this study revealed that African yam bean must have originated from C. cajan and Glycine max before subsequently changing into its present genetic status following evolutionary actions over many years.
GC content is found to be variable with different organisms, the process of which is envisaged to be contributed by variation in selection, mutational bias and biased recombination-associated repair [30]. Also, the GC content of sequences determines their percentage stability in event of evolutionary actions. Guanine and cytosine undergo a specific hydrogen bonding of three bonds making it more thermostable than adenine and thymine bonded by two hydrogen bonds [31]. In our results, the GC content of African yam bean rbcl was similar range with other legumes suggesting that they may all have similar molecular stability. This goes to confirm the result of the phylogenetic grouping of the plants, which showed the possibility common descendant. It can therefore be said that African yam bean is closely related with other legumes used in this study. In terms of the GC content, G. max may be more stable since it had the highest GC content. Also, S. stenocarpa (AYB) may have the exact thermostability properties as V. unguiculata (cowpea) since they both had GC content of 43.36%.
Since all the legumes had similar GC content, it may also be the reason while one type of domain (low complexity) was detected in the protein sequence of the rbcl gene and again, suggesting that AYB and other legumes from fabaeae used in this study are highly similar with similar conserved regions within the rbcl gene. It is important to note that while S. stemecarpa, P. vulgaris, V. argularis and V. unguiculata all had two low complexity domains each, C. cajan and Glycine max had only one low complexity domain. The perculiarity in the number of domains in the rbcl gene may suggest while C. cajan and G. max were grouped into different clade separated from the other legumes [32-42].
The phylogenetic free divided all the legumes into two major cluster, however, African yam bean was sub-grouped with Phaseolus vulgaris, Vigna argularis and Vigna unguiculata. This suggest that the evolutionary charges that have occurred in the sequences of the legumes are similar. Maternal lineage analysis in this study revealed that African yam bean must have originated from C. cajan and Glycine max before subsequently changing into its present genetic status following evolutionary actions over many years. CG content of African yam bean rbcl was similar range with other legumes suggesting that they may all have similar molecular stability. Following the findings of the present study, legumes with the fabaceae family used in this study had high similarity in their rbcL gene sequences.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref]
Citation: Osuagwu AN, Edem UL, Bassey OP, Ije EC (2024) Evaluation of rbcL Gene in African Yam Bean (Sphenostylis stenocarpa Hochst Ex. A Rich Harms) and Related Legumes Using in Silico Approach. J Proteomics Bioinform. 17:660
Received: 02-Feb-2024, Manuscript No. JPB-23-28309; Editor assigned: 05-Feb-2024, Pre QC No. JPB-23-28309 (PQ); Reviewed: 19-Feb-2024, QC No. JPB-23-28309; Revised: 26-Feb-2024, Manuscript No. JPB-23-28309 (R); Published: 04-Mar-2024 , DOI: 10.35248/0974-276X.24.17.660
Copyright: © 2024 Osuagwu AN, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.