Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2023)Volume 16, Issue 2
This research was conducted to assess variation in secondary and tertiary protein structure of rbcL gene of African Yam Bean (Sphenostylis sternocarpa), Phaseolus vulgaris, Vigna unguiculata, Vigna angularis, Glycine max and Cajanus cajan. The rbcL genes of the legume was downloaded from National Center for Biotechnology Information (NCBI) and subjected to multiple sequence alignment on Molecular Evolutionary Genetics Analysis (MEGA). Secondary and tertiary protein structure variations were determined using Goriv and Phyre 2 online computational analysis tools respectively. The phylogenetic relationship among the legumes divided them into two groups. Secondary protein structure subunits were similar among the legumes. However, the folding of the tertiary protein structure was different among the plants suggesting evolutionary actions on the rbcL gene overtime.
Protein structure; Phylogeny; rbcL gene; Subunits
Legumes constitute a large plant family that presents humans with a treasure trove of resources for a variety of uses. Globally, legumes provide important sources of protein, oil, mineral nutrients, and nutritionally important natural products. Grain legume species, including African Yam Bean (Sphenostylis stenocarpa) and adzuki beans (Vigna angularis) are widely used as food and animal feed, cowpea (Vigna unguiculate), pigeon pea (Cajanus cajan), common beans (Phaseolus vulgaris) account for over 33% of human dietary protein, Refined oils, such as soybeans (Glycine max) oil, have industrial applications in paint, diesel fuel, electrical insulation and solvents. Legumes also accumulate phytochemicals, including, which impact human health through pharmaceutical use and as dietary supplements (Dixon and Sumner). An important feature of legumes is their ability to obtain nutrients via symbiosis with soil microbes. The formation of nitrogen-fixing nodules via interaction with bacteria collectively known as rhizobia is virtually unique to legumes, although some species in eight families of the eurosid I clade of dicots can form nodules in association with nitrogen-fixing actinomycetes [1].
The importance attached to the leguminous family cannot be overemphasized, especially in mitigating protein deficiency in the rural population, which is more than 60% of the entire population in most sub-Saharan African countries, including Nigeria.
Unfortunately, there is a decrease in agro-biodiversity of these species in many parts of Nigeria probably due to lack of awareness of the potentials of these crops, poor methods of propagation, processing, marketing and consumption of these crops. These crops have also undergone little or no genetic improvement to boost its agronomic and nutritional qualities. Thus, an understanding of the genes existing in these crops and the proteins which they encode as well as their structure can consequently lead to the genetic improvement of these legumes. The process of identification and characterization of existing germplasm of crops will entail carrying out a robust genetic diversity analyses using molecular/sequence data information. This can be achieved in-silico by the application and utilization of sequenced data.
Presently, many chloroplast, mitochondrial and nuclear genes have been utilized for studying and understanding sequence variations and evolutionary trends at the genus level. One of such Chloroplast gene is Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Large. rbcL gene has been reported to be useful for the study of phylogenetic relationship of flowering plants at the species and generic level. The RuBisCO large subunit (rbcL) plastid marker was commonly used to study unknown taxonomic position of species to elucidate taxonomic connections between different species.
The ribulose biphosphate carboxylase (rbcL) sequence method has been extensively used in studies of evolution, phylogeny, biogeography, population genetics, and systematics because it can be readily copied and not strikingly different for related species [2]. The sequence of rbcL has been recorded in many studies and it is clear that this marker has great potential and benefit in terms of studying the genetic variations of the natural populations [3]. It has also been demonstrated that rbcL primers are useful for inter- and intraspecific evolutionary studies of plants [4]. The discriminatory functions of rbcL gene are executed by the proteins encoded on it.
Proteins are the building blocks of all cells in our bodies and in all living creatures of all kingdoms. Although the information necessary for life to go on is encoded by the DNA molecule, the dynamic process of life maintenance, replication, defense and reproduction are carried out by proteins. Protein structures are responsible for the cell integrity and cell overall shape; their functions vary from composing the cytoskeleton to assembling transmembrane ion channels, structure essential for cell osmolarity and even for synaptic information flux [5]. Not only the DNA molecule cannot replicate itself without protein machinery such as the transcription complex or transcription bubble, but also the mitosis and meiosis events of cell duplication and gamete production cannot go on without proteins performing crossing-over events and chromosome segregation [6].
An understanding of the secondary and tertiary structures of a protein will give us detail information of its biological activity. This study aimed to use bioinformatics tools to characterize the rbcL gene based on secondary and tertiary protein structures of some selected legumes.
Despite the numerous benefits of legumes in agricultural productivity and food security, most of them suffer several limitations ranging from the presence of secondary metabolites, lengthy lifecycle, photoperiodic sensitivity and low yields. Up until now, the genetic diversity present in some legumes remains poorly understood. Chloroplast gene markers are considered useful tools to trace demographic history, explore species divergence and identify species. One of such Chloroplast gene markers universally implored in genetic diversity studies is ribulose biosphosphate carboxylase Large (rbcL) gene given its universality, ease of amplification and alignment. The discriminatory functions of rbcL gene are executed by the proteins encoded on it, thus, an understanding of the secondary and tertiary structures of a protein will give us detail information of its biological activity. This research was conducted to assess variation in secondary and tertiary protein structure of rbcL gene of Sphenostylis sternocarpa, Phaseolus vulgaris, Vigna unguiculata, Vigna angularis, Glycine max and Cajanus cajan. The aim of this study is to evaluate secondary and tertiary protein structure variation in rbcL gene of six legumes.
Experimental site
The in silico study was carried out in the bioinformatics laboratory of the Department of genetics and biotechnology, University of Calabar, Calaabar.
Retrieval of nucleotide and amino acid sequences
Nucleotide sequences of African yam bean and five related legumes in the family of Fabaceae were downloaded from gene bank at National Centre for Biotechnology Information. This was done by downloading the FASTA format for the nucleotide sequences. The Genbank accession numbers of the nucleotide sequences were; Sphenostylis stenocarpa (OK254195.1), V. unguiculata (OK104821.1), Vigna angularis (MH391973.1), Cajanus cajan (OP710494.1), Phaseolus vulgaris (MN078233.1) and Glycine max (LC743621.1).
Phylogenetic analysis
The phylogenetic relationship between African yam bean and the selected related legumes was determined using Molecular Evolutionary Genetic Analysis (MEGA 6) software. Nucleotide sequences download from Genbank were aligned using multiple sequence alignment of ClustalW excluding all the gaps. The phylogenetic grouping was based on 1000 bootstrap replicates.
Prediction secondary and tertiary protein structure of the rbcL gene
Amino acid sequences of the rbcL gene were used to predict the secondary protein structure using GORIV online, while the tertiary protein structure (protein 3D structure) of gene was predicted based on the canonical amino acid sequence obtained from NCBI data with phyre2.
Phylogenetic relationship between AYB and related legumes
The phylogenetic relationship among the legumes and African Yam Bean is presented in Figure 1. The phylogenetic tree was divided into two major clusters. Cluster I consisted of two (G.max and C.cajan) while cluster III consisted of S.stenocarpa, P.vulgaris, V.angularis and V.unguiculata. Furthermore the major cluster II was sub-divided into two sub-clusters V.unguiculata and V.angularis occupying the same sub-cluster while P.vulgaris and S.stenocarpa occupied the same sub-clusters.
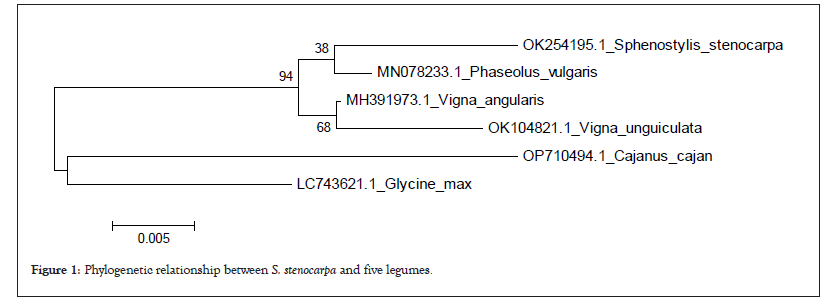
Figure 1: Phylogenetic relationship between S. stenocarpa and five legumes.
Protein structure variation
Table 1 shows percentage of the secondary protein structure subunits in all the legumes used in this study. Alpha helix was not detected in all the protein sequences of the legume plants except in G.max. Extended strand was highest in the G.max (47.45%) while the lowest was in S.stenocarpa and P.vulgaris (43.80%). Again, the percentage of random coil in S.stenocarpa and was the same (56.20%) which was the highest in all the legumes. The tertiary protein structure of rbcL in all the legumes is presented in Figures 2-7. The folding of the tertiary protein structures in all the legumes was different with different proportions of the alpha helix, beta sheet, loops and coils.
| Subunit (%)s | S. stenocarpa | P. vulgaris | V. angularis | V. unguiculata | C. cajan | G. max |
|---|---|---|---|---|---|---|
| Alpha helix | 0 | 0 | 0 | 0 | 0 | 2.92 |
| Extended strand | 43.8 | 43.8 | 46.38 | 47.83 | 45.65 | 47.45 |
| Random coil | 56.2 | 56.2 | 53.62 | 52.17 | 54.35 | 49.64 |
Table 1: Secondary protein structure subunits in rbcl gene of African yam bean and related legumes.

Figure 2: Tertiary protein structure of S. stenocarpa.

Figure 3: Tertiary protein structure of P. vulgaris.
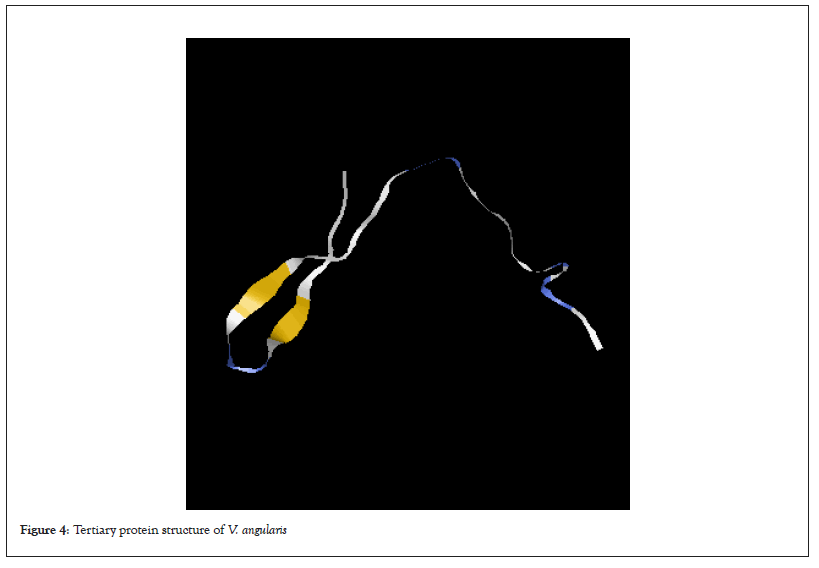
Figure 4: Tertiary protein structure of V. angularis.
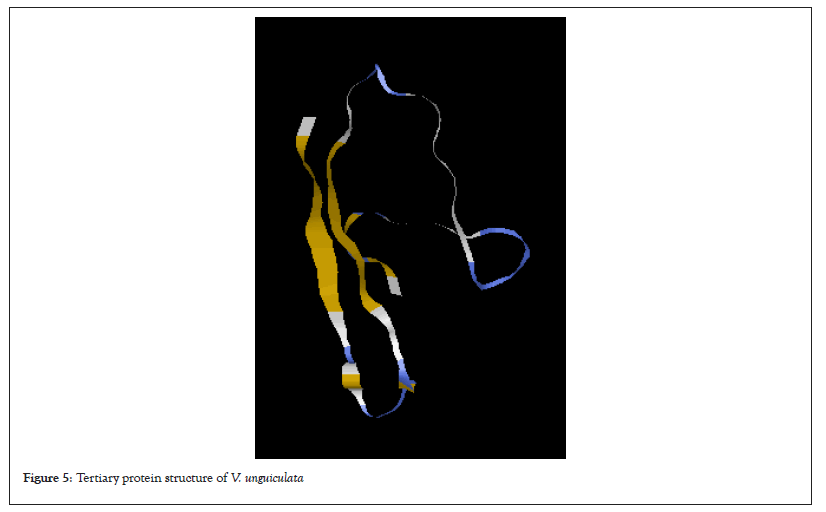
Figure 5:Tertiary protein structure of V. unguiculata.
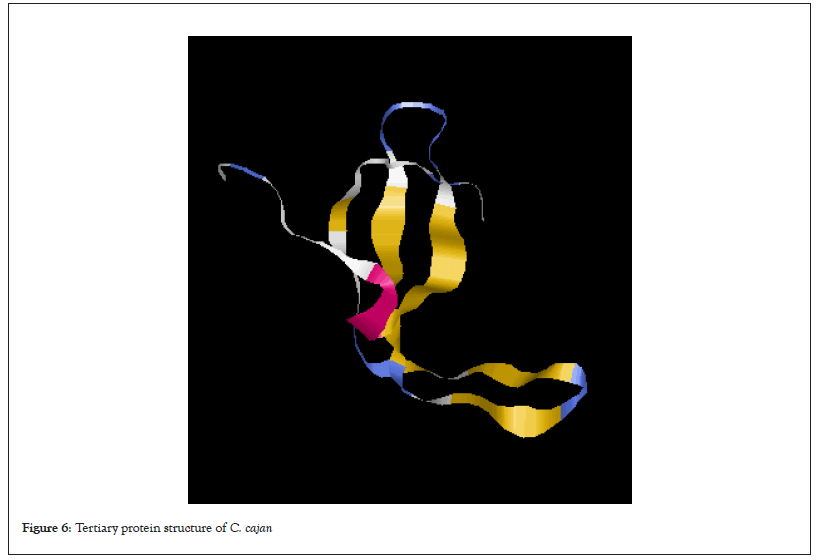
Figure 6: Tertiary protein structure of C. cajan.
Figure 7: Tertiary protein structure of G. max.
The Computation analysis of biological samples have become an integral part of research in recent years opening up a research niche that provides quick answers to many research questions. One of the commonest ways to answer biological questions through Computational studies is to use the phylogenetic methods. A phylogenetic tree is a visual representation of the relationship between different organisms, showing the path through evolutionary time from a common ancestor to different descendants [7].
In our study, the evolutionary relationship of African Yam Bean with related legumes showed that AYB is most related to Phaseolus vulgaris also called common bean although it can be inferred from the tree that all the legumes shared a common ancestor. The relationship shared by the legumes via the phylogenetic tree could suggest their relatedness within their rbcL protein sequences. Therefore, legumes within the same clade in a tree are more likely to have similar protein sequences and structural folding. Molecular phylogenetic uses structure and function of molecules and how they change over time to infer evolutionary relationships. The results of the phylogenetic relationship among the legumes is similar to earlier to earlier findings of who also reported two major clusters among legume family.
Protein structure analysis revealed that alpha helix was not present in all the rbcL sequences of the legumes except in G.max, which had infinitesimal percentage. On the other hand, the percentage of extended strand and the random coil were higher. The variation exhibited by the protein subunit among the studied legume is a confirmation although they may share common ancestor, there has been evolutionary factors such as mutation and other environmental factors that have to modify their genome to give them genetic peculiarity. According to Weinberg 1994, the structure of protein could reveal its function and evolutionary history. Alpha helixes and beta sheets are the two most common secondary structural elements in protein structure although beta turns and omega loops can occur. Secondary structure elements spontaneously form as an intermediate before proteins fold into its three Dimensional (3D) tertiary structure.
Variation in proteins have very large number of diverse effects on their sequence, structure, stability, activity, abundance as well as other characteristics [8,9]. Factors such a pH, temperature, oxidizing/reducing agents are potential causes of protein sequence variations. These could suggest that the variation observed in the secondary and tertiary protein structures of AYB and other legumes maybe as a result of the influence of some of these factors.
This Although the secondary protein subunit indicated high similarity among the rbcl gene of African Yam Bean and other legumes in this study, variations observed in the folding’s of the 3D structure of the protein sequences suggest evolvement of the Gene among the legume plants overtime. Although many attempts to unravel protein stability and the relationship between sequences of protein and its three-dimensional structure focuses on the properties of the native folded proteins important insight can also from studies of denatured and partly folded proteins. Also the characterization of protein at such non-native states can help in understanding protein folding, transportation across membranes and protein turnover in cell. This may also explain the nature of the folding’s of protein sequences of African yam bean (AYB) and other related legumes of AYB and other related legumes observed in this research.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Edem UL, Osuagwu AN (2023) Evaluation of Secondary and Tertiary Protein Structure Variation in rbcl Gene of Selected Legumes using Computational Approach. J Proteomics Bioinform.16:642.
Received: 03-May-2023, Manuscript No. JPB-23-23872; Editor assigned: 05-May-2023, Pre QC No. JPB-23-23872 (PQ); Reviewed: 19-May-2023, QC No. JPB-23-23872; Revised: 26-May-2023, Manuscript No. JPB-23-23872 (R); Published: 06-Jun-2023 , DOI: 10.35248/0974-276X.23.16.642
Copyright: © 2023 Edem UL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : no