
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2021)Volume 11, Issue 6
Background: Noninvasive Prenatal Testing (NIPT) is based on second-generation genomic sequencing technology to scan cell-free fetal DNA originating from the placenta in maternal plasma. As the depth of sequencing increases, it can be used to focus on chromosomal aneuploidies, Copy Number Variants (CNVs), and monogenic diseases. It can significantly improve the accuracy of prenatal screening and reduces the number of invasive testing.
Methods: In this study, we retrospectively analyzed 16128 naturally conceived singleton pregnancies, which underwent expanded NIPT to calculate the True Positive Rate (TPR) of chromosomal aneuploidies and CNVs, and analyzed the potential influence of maternal Sex Chromosome Abnormalities (SCAs) and maternal CNVs on expanded NIPT results.
Results: After invasive prenatal diagnosis and follow-up, 103 pregnancies were found to be true-positive, including 73 cases of chromosomal abnormalities and 30 cases of CNVs. The TPR of T21 was 84.62%, T18 was 50.00%, T13 was 22.22%, SCA was 34.06%, and CNVs was 40.28%. The false negative rate and the sensitivity of expanded NIPT for fetal trisomy’s 2,118 and 13 was found to be 0.0062% and 99.99%, respectively.
Conclusion: Expanded NIPT showed good performance in detecting diseases of chromosomal abnormalities and CNVs, and was not easy to miss true positive, but there would be relatively high false positive rate and maternal SCAs and CNVs may confuse some NIPT results. Therefore objectively understand its advantages, limitations and indications, as well as clinical consultation before and after the NIPT are critical.
Noninvasive prenatal testing; Copy number variants; Sex chromosome abnormalities; Maternal copy number variants; Advanced maternal age
NIPT: Noninvasive Prenatal Testing; CNVs: Copy Number Variants; TPR: The True Positive Rate; FPR: The False Positive Rate; SCAs: Sex Chromosome Abnormalities; cffDNA: The cell-free fetal DNA; MMS: Microdeletion/Microduplication Syndromes; PPV: Positive Predictive Value; MCNV: Maternal Copy Number Variants; pCNVs: Pathogenic Copy Number Variations; CPM: Confined Placental Mosaicism; DMD: Duchenne Muscular Dystrophy; IVF: In Vitro Fertilization.
The clinical use of Noninvasive Prenatal Testing (NIPT) using maternal plasma to detect fetal genetic material was made possible by the discovery of cell-free fetal DNA (cffDNA) in the maternal circulation in 1997 [1] and the development of next-generation sequencing in 2008 [2]. This technological innovation significantly reduces the number of invasive tests, and increases the efficiency of invasive prenatal screening [3]. A large number of clinical studies have shown that NIPT has a high sensitivity and specificity for diseases of chromosomal aneuploidy. The true-positive rates range of T21 (Down's syndrome) was 65%-95%, T18 (Edward's syndrome) was 47%-85%, and T13 (Patau syndrome) was 12–62% [4-6].
As the depth of sequencing increases and the calculation methods change, the focus is on aneuploidies, Copy Number Variants (CNVs), and monogenic diseases. CNVs cause microdeletion/Microduplication Syndromes (MMS), which are unlikely to be detected by ultrasound examination and have a much higher incidence than Down syndrome [7], accounting for 1%-2% of newborn congenital abnormalities [8]. Studies have suggested that expanded NIPT yielded high Positive Predictive Values (PPV) for common aneuploidies and DiGeorge syndrome, and moderate PPVs for other MMS [9]. However, the rate of false positive and false negative results makes the implementation of the expanded NIPT more challenging, therefore requiring validation in clinical practice.
In this retrospective study, we analyzed 16128 patients with naturally conceived singleton pregnancies using expanded NIPT and analyzed the performance of expanded NIPT as a screening test for fetal aneuploidies and CNVs. We also calculated the influence of maternal age, Sex Chromosome Abnormalities (SCAs), and Maternal Copy Number Variants (MCNV) on the positive rate of fetal aneuploidies and CNVs.
Patients
This study was designed as a retrospective study, and the inclusion criteria were as follows: 1) single pregnancy, and 2) natural conception. The exclusion criteria were as follows: 1) multiple pregnancy, 2) conception through IVF (in vitro fertilization), 3)received immunotherapy within 4 weeks of NIPT, and 5) first NIPT test failed. According to the above criteria, a total of 16128 pregnant women were recruited from February 2017 to December 2020. Venous blood samples were collected from the Gansu Province Maternal and Child Health Care Hospital in Lanzhou, China. All the participants purchased Taikang insurance under a specific expanded NIPT insurance scheme covering the standard and expanded test range. Informed written consent was obtained from all participants who agreed to receive expanded NIPT. Pregnancies were divided into the following groups: Fetal structural abnormalities by ultrasound (including NT ≥ 3 mm), high risk of serological screening (T21>1/270, T18>1/350), advanced maternal age ( ≥ 35 years), critical risk of serological screening (T21 1/270 to 1/1000, T18 1/350 to 1/1000), No serology screening, and no clinical indications (low risk of serological screening, no abnormalities on ultrasound and no advanced maternal age). The study was approved by the hospital ethics committee, and all pregnancy signed an informed consent form.
Library construction and DNA sequencing
We collected 8 to 10 mL of whole blood samples in special tubes (Streck, USA). Plasma separation was performed at 4°C within 72 h of blood sample collection. Afterwards, cell-free DNA extraction and purification, library construction, quality control, quantification, addition of sequence tags, and pooling were performed according to the fetal chromosome aneuploidies (T21/ T18/T13) test kit (Berry Genomics, China). Finally, the samples were sequenced on the NextSeq CN500 platform (Illumina, USA). Sequencing reads were mapped to the human reference genome (GRCh37/hg19). Sequencing and analysis were performed as previously described [9].
Prenatal diagnosis
Each participant received counselling after expanded NIPT screening. Positive expanded NIPT individuals were recommended to receive invasive prenatal diagnosis. Invasive prenatal diagnosis and follow-up results were used as the gold standard to calculate the true positive case. Whole chromosomal aneuploidies were confirmed by karyotyping and CNVs were confirmed by CNV-Seq. The pathogenicity of CNVs was evaluated following the ACMG guidelines.
Peripheral blood test
Study participants with Sex Chromosomal Abnormalities (SCAs) detected by expanded NIPT were also recommended to receive a peripheral blood FISH test. Participants with CNVs detected by expanded NIPT were also recommended to receive Parents' peripheral blood CNV-Seq. The pathogenicity of the CNVs was evaluated following the ACMG guidelines.
Pregnancy characteristics
A total of 16128 naturally conceived singleton pregnancies were included in this study.
The maternal age ranged from 15 to 55 years-old and the pregnancy gestations ranged from 11+0 to 32+6 weeks. Of all the participants in the study, 2735 had a history of more than two spontaneous abortions or pregnancies have been pregnant or birth defect, that called had a history of adverse pregnancy and childbirth (16.96%). Among the 16128 participants who underwent expanded NIPT, 1201 (7.45%) showed fetal structural abnormalities by B-ultrasound (including NT ≥ 3 mm), 1785 (11.07%) showed a high risk of serological screening, 5143 (31.89%) showed a critical risk of serological screening, 4889 (30.31%) had advanced maternal age (age ≥ 35), 2295(14.23%) showed no serology screening, and 815 (5.05%) had no clinical indications in Table 1.
| Maternal age at NIPT (years) | No./N=16128 | Rate (100%) |
|---|---|---|
| <30 | 6062 | 37.59 |
| 30-34 | 4916 | 30.48 |
| 35-40 | 4858 | 30.12 |
| ≥ 41 | 292 | 1.81 |
| Range | 15-55 | |
| Gestational age at NIPT (weeks) | No. | Rate (100%) |
| 11-15+6 | 4612 | 28.6 |
| 16-22+6 | 9782 | 60.65 |
| 23-32+6 | 1734 | 10.75 |
| Range | 11-32+6 | |
| History of adverse pregnancy and childbirth | No./N=16128 | Rate (100%) |
| Yes | 2735 | 16.96 |
| No | 13393 | 83.04 |
| Clinical features | No./N=16128 | Rate (100%) |
| Fetal structural abnormalities by B-ultrasound | 1201 | 7.45 |
| High risk of serological screening | 1785 | 11.07 |
| Critical risk of serological screening | 5143 | 31.89 |
| Advanced maternal age ( ≥ 35 years) | 4889 | 30.31 |
| No serology screening | 2295 | 14.23 |
| No clinical indications* | 815 | 5.05 |
Note: *No clinical indications, low risk of serological screening, no abnormalities on ultrasound and no advance maternal age.
Table 1: Maternal characteristics and gestational age.
The performance of expanded NIPT
Of the 16128 participants that underwent expanded NIPT, 287 abnormal results were detected, and diagnostic testing by karyotyping and CNV-Seq was used to verify the abnormal results. Among the 287 cases, 60 refused prenatal diagnosis and the remaining 227 cases were verified and followed up with the following results: 103 true positives (33 cases of T21, 7 of T18, 2 of T13, 31 of SCAs, 30 of CNVs); 124 false positives (6 cases of T21, 7 of T18, 7 of T13, 60 of SCA, 44 of CNVs); and one false negative (T21). Moreover, the True Positive Rate (TPR) and the False Positive Rate (FPR) for each test was assessed. For trisomy 21 (T21), the TPR was 84.62% (95%CI, 73.30%-95.94%), the FPR was 15.38%, for trisomy 18 (T18), the TPR was 50.00% (95%CI, 23.81%-76.19%), the FPR was 50.00%. For trisomy 13 (T13), the TPR was 22.22% (95% CI, 4.94%-49.38%), the FPR was 77.78%. For SCAs, the TPR was 34.06% (95% CI, 24.32%-43.79%), the FPR was 65.94%. For CNVs, the TPR was 36.25% (95%CI, 25.30%-47.20%), the FPR was 63.75% in Table 2.
| NIPT | T21 | T18 | T13 | SCAs | CNVs |
|---|---|---|---|---|---|
| Positive | 46 | 18 | 10 | 121 | 98 |
| Unverified | 7 | 4 | 1 | 30 | 18 |
| TP | 33 | 7 | 2 | 31 | 29 |
| FP | 6 | 7 | 7 | 60 | 51 |
| TPR | 84.62 | 50 | 22.22 | 34.06 | 36.25 |
| FPR | 15.38 | 50 | 77.78 | 65.94 | 63.75 |
Table 2: Performance of expanded NIPT.
TPRs of chromosomal aneuploidies according to pregnancy characteristics
As shown in Table 3, different pregnancy characteristics correspond to different TPRs. The total TPR of T21 was 84.62% (95%CI, 73.30%-95.94%). In both the high risk of serological screening group and the no serology screening group, the TPRs of T21 were the highest at 100%, while the TPRs of T21 in the advanced maternal age group, the B-Ultrasound indicated abnormalities group, and the critical risk of serological screening group were 92.86%, 83.33% and 66.67%, respectively. The total TPR of SCAs was 34.06% (95% CI, 27.06%-41.06%), with the highest being 50.00% in the ultrasound indicated abnormalities. The TPRs of T18 in the high risk of serological screening group, the critical risk of serological screening group, advanced maternal age group, and the no serology screening group were 16.67%, 30.77%, 39.28%, and 28.57%, respectively. Among the 153 positive cases which underwent invasive prenatal diagnosis, the number of true positive cases was 73, the number of false positive case was 80, and the overall TPR of aneuploidies was 47.71%. The TPRs of aneuploidies in the B-Ultrasound indicated abnormalities group, the high risk of serological screening group, the Critical risk of serological screening group, the advanced maternal age group, and the no serology screening group were 55.56%, 36.84%, 40.90%, 58.00%, and 35.00%, respectively.
| Clinical features | T21 | T18 | T13 | SCAs | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FP | TPR (%) | TP | FP | TPR (%) | TP | FP | TPR (%) | TP | FP | TPR (%) | TP | FP | TRP (%) | |
| Ultrasound indicated abnormalities | 5 | 1 | 83.33 | 2 | 5 | 5 | 50.00 | 10 | 8 | 55.56 | |||||
| High risk of serological screening | 4 | 100 | 1 | 1 | 1 | 50.00 | 2 | 10 | 16.67 | 7 | 12 | 36.84 | |||
| Critical risk of serological screening | 8 | 4 | 66.67 | 1 | 2 | 33.33 | 1 | 2 | 33.33 | 8 | 18 | 30.77 | 18 | 26 | 40.90 |
| Advanced maternal age ( ≥ 35) | 13 | 1 | 92.86 | 5 | 2 | 71.43 | 1 | 11 | 17 | 39.28 | 29 | 21 | 58.00 | ||
| No serology screening | 3 | 100 | 2 | 1 | 4 | 10 | 28.57 | 7 | 13 | 35.00 | |||||
| No clinical indications | / | / | 1 | / | / | / | 1 | / | 2 | / | |||||
| Total | 33 | 6 | 84.62 | 7 | 7 | 50.00 | 2 | 7 | 22.22 | 31 | 60 | 34.06 | 73 | 80 | 47.71 |
Table 3: TPRs of chromosomal aneuploidies according to pregnancy characteristics.
The influence of maternal age on the positive rate of fetal aneuploidies and CNVs
As shown in Figure 1, we divided the study participants by maternal age into four groups to analyze the influence of maternal age on the positive rate of fetal aneuploidies and CNVs. The following positive rates were determined for the different maternal age groups: the <30 years group was 0.36% (95% CI, 0.21%-0.51%), the positive rate of the 30-34 years group was 0.47% (95% CI, 0.29%-0.66%), the 35-40 years group was 0.66% (95% CI, 0.43%-0.89%), the ≥ 41 years group was 2.05% (95% CI, 0.42%-3.68%). A Chi-square test was used to analyze the significance of the differences between the different groups.
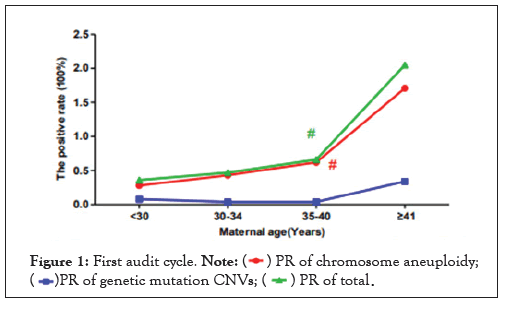
Figure 1: First audit cycle. Note:
For chromosome aneuploidies, the positive rate of the advance maternal age group (the 35-40 group and the ≥ 41 group) was higher than the <35 group (<30 group and 30-34 group), and the difference was statistically significant (χ2=8.651, p=0.003<0.05). For CNVs, the difference between the advance maternal age group (35-40 group and ≥ 41 group) and the <35 years group (<30 group and 30-34 group) was not statistically significant (χ2=0.000, p=1.000>0.05). The total positive rate increased with maternal age, and the positive rate of the advance maternal age group (35-40 group and ≥ 41 group) was higher than the <35 group (<30 group and 30-34 group), and the difference was statistically significant (χ2=4.409, p=0.036<0.05).
The potential influence of parental CNVs on fetal CNVs
Besides T21 T18 and T13, we analyzed other chromosomal aneuploidies and CNVs among the 16128 samples. A total of 98(0.61%) cases were detected to have abnormal CNVs results, 80 cases underwent invasive prenatal diagnosis, while 18 patients refused amniotic fluid puncture. Of the 80 cases that underwent invasive prenatal diagnosis, 29(36.25%) of them were true positives (Table 4), 51(63.75%) cases were false positives (including nine false positive cases where abnormal results were detected in the mother's peripheral blood, while the fetal amniotic fluid was normal). Among the 29 cases where abnormal results were detected in the fetal amniotic fluid, 27 cases underwent Parental peripheral blood verification. Additionally, among the 27 cases, 13(48.15%) cases occurred because of genetic mutations, while 14(51.85%) cases were inherited from parents. Among the 14 cases, 13 cases were inherited from the mother and 1 case was inherited from the father. Among the 29 cases where abnormal results were detected in the fetal amniotic fluid, 9(31.03%) cases were identified as syndrome diseases or pathogenicity, and 15(51.727%) cases had unknown pathogenicity, and 5(17.24%) cases were benign, according to the ACMG guidelines.
| Case | MG | NIPT results | Prenatal diagnosis | Parents' CNV-seq | |
|---|---|---|---|---|---|
| CNV-seq | karyotype | ||||
| A. Genetic mutations | |||||
| 1 | 32 | 7p22.3-p11.2 dup 55.6Mb,7q11.21-q36.3 dup 92.3Mb | 47, XN+7[10%]/46, XN [90%] | 46, XN | normal |
| 2 | 27 | 7p22.3-p11.2 dup 55.6Mb,7q11.21-q36.3 dup 92.3Mb | 47, XN+7(40%)/46, XN (60%) | 47, XN+7[40%] /46, XN [60%] |
normal |
| 3 | 42 | 7q36.2-q36.3 dup4.3Mb, 14q11.2-q21.3 dup 27.4Mb | 7q36.2-q36.3 dup4.44Mb, 14q11.2-q21.3 dup 29.06Mb | 46, XN, dup (14) (q11.2-q21.3) | normal |
| 4 | 25 | 10q.24.2-q26.3 dup 35.8Mb | 10q24.1-q26.3 dup 37.2Mb | 46, XN, dup (10) (q24.1-q26.3) | normal |
| 5 | 27 | 12p13.33-p11.1 dup 34.3Mb | 12p13.33-p11.1 dup34.7Mb | 46, XN, dup (12) (p13.3-p11.1) | normal |
| 6 | 34 | 13q33.3-q34del4.2mb, | 13q33.3-q34del4.86Mb, | 46, XN | normal |
| 7 | 27 | 18p11.32-p11.21 del 13.5 | 18p11.32-p11.21del 14.86Mb | 46, XN | normal |
| 8 | 38 | 22q11 deletion syndrome | 22q11.21 del 2.58Mb | 46, XN | normal |
| 9 | 30 | 6p24.3-p22.3 del 5.2Mb | 6p24.2-p22.3 del 5.12Mb | 46, XN | normal |
| 10 | 28 | 6q23.3-q24.1 dup 2.0Mb | 6q23.3-q24.1 dup 2.88Mb | 46, XN | normal |
| 11 | 29 | 10q22.3-q23.1 del 4.5Mb | 10q22.3-q23.1 del 4.46Mb | 46, XN | normal |
| 12 | 24 | 4q35.2 dup 2.0Mb | 4q35.2 dup 1.92Mb | 45, XN, rob (13; 14) (q10; q10) | normal |
| 13 | 36 | 8p23.1 dup 3.8Mb | 8p23.1 dup 3.76Mb | 46, XN | normal |
| B. Inherited from parents | |||||
| 14 | 27 | 22q11.21 dup 2.4Mb | 22q11 dup 2.5Mb | 46, XN | 22q11 duplication syndrome (M) |
| 15 | 35 | 16p13.11-p12.3 dup 2.7Mb | 16p13.11-p12.3 dup 2.64Mb | 46, XN | 16p13.11-p12.3 dup 2.64Mb (M) |
| 16 | 32 | 16q21 del 4.0Mb | 16q21del3.98Mb | 46, XN | 16q21 del 3.98Mb (M) |
| 17 | 24 | 1p36.33-p36.32 dup 2.4Mb | 1p36.33-36.32 dup 2.26Mb | 46, XN | 1p36.33-36.32 dup 2.38Mb (M) |
| 18 | 32 | 2p22.3 dup 2.3Mb | 2p22.3 dup 2.20Mb | 46, XN | 2p22.3 dup 2.20Mb (M) |
| 19 | 32 | 3q11.1-q11.2 dup3.2Mb | 3q11.1-q11.2 dup 3.06Mb | 46, XN, inv (9) | 3q11.1-q11.2 dup 3.10Mb(M) |
| 20 | 28 | 4q34.3 dup 2.0Mb | 4q34.3 dup 1.66Mb | 46, XN | 4q34.3 dup 1.62Mb (M) |
| 21 | 26 | 4q34.3 dup 2.2Mb | 4q34.3 dup 1.62Mb | 46, XN | 4q34.3 dup 1.72Mb (M) |
| 22 | 26 | 4q34.3 dup 2.4Mb | 4q34.3 dup 2.28Mb | 46, XN | 4q34.3 dup 2.22Mb (M) |
| 23 | 37 | 7p21.3 dup 3.1Mb | 7p21.3 dup 0.7Mb | 46, XN | 7p21.3 dup 1.4Mb (M) |
| 24 | 22 | 8p23.2 dup 2.3Mb | 8p23.2 dup 2.22Mb | 46, XN | 8p23.2 dup 2.22Mb (M) |
| 25 | 33 | 8p23.2 dup 2.6Mb | 8p23.2 dup 2.26Mb | 46, XN | 8p23.2 dup 2.26Mb (M) |
| 26 | 34 | 8q24.21-q24.22 dup 2.0Mb | 8q24.21-q24.22 dup 1.3Mb | 46, XN | 8q24.21-q24.22 dup 1.3Mb (M) |
| 27 | 23 | 10q21.1 del 3.1Mb | 10q21.1 del3.04Mb | 46, XN | 10q21.1 del3.04Mb (F) |
| C. Parents' CNVs Unverified | |||||
| 28 | 24 | 3q11.2 dup 2.6Mb | 3q11.1-q11.2 dup 3.16Mb | / | |
| 29 | 25 | 3p12.3-p12.2 dup 2.5Mb | 3p12.3-p12.2 dup 2.5Mb | / | |
Table 4: The true positive of CNVs.
Potential influence of maternal SCAs and CNVs on expanded NIPT
Among the results of expanded NIPT, there were eight cases where we detected normal results in the fetal amniotic fluid, but the FISH analysis showed SCAs in the maternal peripheral blood (Table 5A). Of the 60 false positive cases of fetal SCAs, maternal SCAs accounted for 13.33%. Among expanded NIPT, there were 9 cases where we detected normal results in the fetal amniotic fluid, but the CNV-seq showed abnormal results in the maternal peripheral blood (Table 5B).
| Case | NIPT results | prenatal diagnosis | Maternal peripheral blood |
|---|---|---|---|
| A. Maternal potential influence of SCAs | |||
| 1 | ChrX- | 46, XN | 46, XX [68%]/47, XXX [32%] |
| 2 | ChrX+ | 46, XN | 45, X0 [26%]/46, XX [74%] |
| 3 | ChrX- | 46, XN | 45, X0 [13%]/46, XX [87%] |
| 4 | ChrX+(Y) | 46, XN | 47, XXX |
| 5 | ChrX- | 46, XN | 45, X0 [10%]/46, XX [90%] |
| 6 | ChrX+(Y) | 46, XN | 46, XX [33%]/47, XXX [67%] |
| 7 | ChrX- | 46, XN | 45, X0 [30%]/46, XX [70%] |
| 8 | ChrX- | 46, XN | 45, X0 [20%]/46, XX [80%] |
| B. Maternal potential influence of copy number variations | |||
| 1 | 16p13.11-p12.3 dup 2.7Mb | 46, XN | 16p13.11-p12.3 dup 2.5Mb |
| 2 | 1q21.1-q21.2 dup 2.0Mb | 46, XN | 1q21.1 recurrent micro-duplication |
| 3 | 1q43 dup 2.2Mb | 46, XN | 1q43 dup 0.34Mb |
| 4 | 2p12-p11.2 dup 2.1Mb | 46, XN | 2p12 dup 1.1Mb |
| 5 | 4p13-p12 del 2.5Mb | 46, XN | 4p13-p12 del 2.34Mb |
| 6 | 5q34 del 3.6Mb | 46, XN | 5q34 del 3.5Mb |
| 7 | 6p21.33-p21.32 dup 2.6Mb | 46, XN | 6p21.32 dup 0.26Mb |
| 8 | 8p23.2 dup 2.3Mb | 46, XN | 8p23.2 dup 2.26Mb |
| 9 | 8q21.13 dup 2.1Mb | 46, XN | 8q21.13 dup 1.9Mb |
Table 5: The influence of maternal SCAs and CNVs on expanded NIPT results.
Compared to traditional prenatal screening methods based on serological screening and ultrasound screening to assess fetal chromosomal abnormalities, NIPT is a more accurate prenatal screening tool. The detection rate of chromosomal abnormalities of traditional prenatal screening is 50%-95% [10], while the sensitivity and specificity of NIPT for fetal trisomy’s 21, 18, and 13 are higher than 99% [11]. In our study, in addition to invasive prenatal diagnosis, we also conducted follow-up. Besides the unverified cases, we found 20 false positive cases and one false negative case of T21. The false positive rate and the specificity of expanded NIPT for fetal trisomy’s 21, 18, and 13 was found to be 0.12% and 99.88%, respectively. The false negative rate and the sensitivity of expanded NIPT for fetal trisomy’s 21, 18, and 13 was found to be 0.0062% and 99.99%, respectively. Our results also show that the TPR of T21 was 84.62% (95% CI, 73.30%- 95.94%), the TPR of T18 was 50.00% (95% CI, 23.81%-76.19%), the TPR of T13 was 22.22% (95% CI, 4.94%-49.38%), and the TPR of CNVs was 40.54% (95% CI, 29.35%-51.73%), it was higher than that of T13 and close to T18. Expanded NIPT is not only more accurate but also avoids unnecessary invasive prenatal diagnosis methods which may result in approximately 0.1%–0.3% procedure-related pregnancy loss [12], and more and more pregnant women are willing to choose expanded NIPT [13]. With the deepening of sequencing, the expanded NIPT, which detects aneuploidies and genome-wide MMS caused by CNVs, has become available. Studies have shown that about 80% of pregnant couples in the Netherlands are willing to choose whole genome testing instead of common trisomies [14].
According to China’s guidelines for NIPT published on October 27, 2016, NIPT should be used with caution for pregnant women older than 35 and for patients with a high risk of serological screening. However, our findings reveal that many of them opted for NIPT to avoid invasive prenatal diagnosis. This observation is also supported by the study by Tian et al. [15]. Among 16128 pregnant women, 4889 (30.31%) of them were older than 35, and 1785 (11.07%) of them showed a high risk of serological screening. Simultaneously, in the older than 35 groups, the TPR of T21 was 92.86%, the TPR of T18 was 71.43%, and the TPR of SCAs was 39.28%. As such, the expanded NIPT reduced the incidence of invasive procedures. Consistent with previous studies [16], we found that the prevalence of fetal aneuploidies increased with the maternal age. Our study shows that for chromosomal aneuploidies, the incidence of abnormal results tends to increase with the maternal age. Taking 35 as the node, the positive rate of abnormality in advanced maternal age group is higher than the <35 years old group, and the difference is statistically significant. As reported in other studies, the common CNVs are not related to maternal age [17], and our findings suggest that the positive rate of CNVs with the different maternal age groups is trending but not statistically significant.
According to China’s guidelines for NIPT published on October 27, 2016, NIPT should be used with caution for pregnant women older than 35 and for patients with a high risk of serological screening. However, our findings reveal that many of them opted for NIPT to avoid invasive prenatal diagnosis. This observation is also supported by the study by Tian et al. [15]. Among 16128 pregnant women, 4889 (30.31%) of them were older than 35, and 1785 (11.07%) of them showed a high risk of serological screening. Simultaneously, in the older than 35 groups, the TPR of T21 was 92.86%, the TPR of T18 was 71.43%, and the TPR of SCAs was 39.28%. As such, the expanded NIPT reduced the incidence of invasive procedures. Consistent with previous studies [16], we found that the prevalence of fetal aneuploidies increased with the maternal age. Our study shows that for chromosomal aneuploidies, the incidence of abnormal results tends to increase with the maternal age. Taking 35 as the node, the positive rate of abnormality in advanced maternal age group is higher than the <35 years old group, and the difference is statistically significant. As reported in other studies, the common CNVs are not related to maternal age [17], and our findings suggest that the positive rate of CNVs with the different maternal age groups is trending but not statistically significant.
The incidence of birth defects in China is about 5.6% [18]. Chromosomal aberrations account for more than 80% of the genetic causes, including abnormal number of chromosomes, and pathogenic copy number variations (pCNVs, which account for 50%) [19]. So far, more than 300 types of pCNVs have been found to cause chromosome microdeletion/microduplication syndrome, and the comprehensive incidence rate is nearly 1/600 [20]. Therefore, effective prenatal screening and subsequent timely prenatal diagnosis for chromosomal aberrations is critical for reducing the birth defects of live births. Expanded NIPT performance in some CNVs has been thoroughly described. A prospective study which involved a large group of pregnant women showed that expanded NIPT exhibited high performance for the 22q11.2 microdeletion, and moderate-to-low performance for detection of other, genome-wide, segmental imbalances associated with other MMS and some CNVs [9]. In this study, we found 29 (36.25%) true-positive cases of chromosomal microdeletions or microduplications that were validated by CNV-seq. Among the 29 true-positive cases, 13 cases occurred because of genetic mutations. We also found 51 (63.75%) false-positive cases. Among the 51 falsepositive cases, 9(17.65%) cases occurred because of abnormalities in the maternal peripheral blood, consistent with other literature that showed MCNV can potentially contribute to a small but significant number of false-positive fetal trisomies detected by NIPT [21]. NIPT uses cell-free fetal DNA (cffDNA) extracted from maternal plasma, which is a mixture of maternal DNA and a low percentage of fetal DNA. Therefore, chromosomal aneuploidy and CNVs abnormalities of pregnancy have a great influence on NIPT results, making the reliable and accurate detection of aneuploidies or MMS challenging [22]. A study reported that altered maternal X chromosome karyotype and maternal X CNVs contribute to discordant NIPT SCAs results [23]. In this study, we found 31 (34.06%) true-positive cases for SCAs that were validated by karyotype and CNV-seq. We also found eight cases that were detected as normal results in the fetal amniotic fluid, but the FISH test showed SCAs in maternal peripheral blood, which accounts for 13.33% of the false positive SCAs cases. From this data, we can conclude that the pregnancy SCAs and CNVs have a great influence on the accuracy of NIPT results. Apart from pregnancy SCAs or CNVs, low fetal DNA fraction and Confined Placental Mosaicism (CPM) [24] can confound any NIPT results.
Almost all cffDNA in maternal blood comes from placental trophoblast cells [25], however the fetus originates from the inner cell population of the cytotrophoblast, the results of NIPT may not always represent the true condition of the fetal chromosomes, so it is a screening test. Many studies have shown that, compared with traditional screening technologies, expanded NIPT has better sensitivity and accuracy for detecting Chromosome aneuploidy [9,17,26,27], and it is feasibility for detecting fetal CNVs. Our study shows that the false positive rate and the specificity of expanded NIPT for fetal trisomy’s 21, 18, and 13 was found to be 0.12% and 99.88%, respectively. The false negative rate and the sensitivity of expanded NIPT for fetal trisomy’s 21,18, and 13 was found to be 0.0062% and 99.99%, respectively, this is consistent with other report that the incidence of false negative rate is about 0.01% [28]. Our results also show that the TPR of T21 was 84.62%, the TPR of T18 was 50.00%, the TPR of T13 was 22.22%, and the TPR of CNVs was 40.54%. Especially, in the older than 35 groups, the TPR of T21 was 92.86%, the TPR of T18 was 71.43%, and the TPR of SCAs was 39.28%. We can summary that even though NIPT has high accuracy and is not easy to miss true positive, there will be relatively high false positives. Our findings suggest that maternal SCAs and CNVs contribute to a small but significant number of false-positive fetal trisomies and CNVs detected by NIPT. Therefore, to avoid false positive caused by maternal SCAs or CNVs and avoid unnecessary invasive procedures, we recommend that there is a need to develop a new analysis or calculation method to remove the potential pregnancy influence on expanded NIPT results.
As the depth of sequencing increases and calculation methods change, monogenic diseases, such as congenital adrenal hyperplasia, Duchenne Muscular Dystrophy (DMD) and others may also be identified via expanded NIPT [29-31]. However, it remains some defects such as unable to detect chromosome structural variations, unable to avoid false positives and false negatives, unable to remove the influence of maternal abnormalities until now; these defects will make such a high rate of women being unsettled after the test [32]. Therefore complete informed consent, clinical consultation before and after the NIPT, objectively understand its advantages, limitations and indications, is the most effective way to solve the current clinical application of NIPT [33].
Our study concludes that expanded NIPT shows good performance in detecting diseases of chromosomal aneuploidy and CNVs, but it remains some defects such as unable to avoid false positives and false negatives and unable to remove the influence of maternal abnormalities until now. These defects may make such a high rate of women being unsettled after the test. Therefore, in order to improve the accuracy of detection, there remains a need to reduce the false positive and false negative, in order to reduce pregnancy unsettlement after the test, objectively understand its advantages, limitations and indications, as well as clinical consultation before and after the NIPT are critical.
Ethics approval and consent to participate
This study was approved by the Gansu Province Maternal and Child Health Care Hospital ethics committee
Not applicable
Citation: He J, Feng X, Wang X, Zhang Q, Zheng L, Lin P, et al. (2021) Expanded Noninvasive Prenatal Testing for Chromosomal Aneuploidies and Copy Number Variants in a Cohort of 16128 Single Pregnancies. J Clin Trials. 11:479.
Received: 24-Aug-2021 Accepted: 07-Sep-2021 Published: 14-Sep-2021 , DOI: 10.35248/2167-0870.21.11.479
Copyright: © 2021 He J, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.