Enzyme Engineering
Open Access
ISSN: 2329-6674
ISSN: 2329-6674
Short Communication - (2022)Volume 11, Issue 4
Barbiturates are the first line drugs for treatment of epilepsy for adults and children in the developing world because of its low cost and proven effectiveness. Due to their adverse effects, no effective management of toxicity and difficulty in determining correct dosage, benzodiazepines are preferred over them. It is also used as a recreational drug, thereby contributing to cases of overdose. My present study was to find the inhibitors of barbiturates by in-silico molecular docking. This approach can highlight only competitive inhibitors as molecules in-silico are rigid and do not produce conformational changes in structure by allosteric binding. 450 FDA-approved drugs were docked to active site of barbiturate of Gleobacter Ligand-Gated Ion Channel (GLIC). Drug interactions were visualized, literature search was done to bring out the final results. Tolazamide, an oral anti-diabetic drug and 5-methyltetrahydrofolate, active metabolite of folic acid produced desired results. These results should be used clinically for validation.
Barbiturate; Toxicity; Tolazamide; 5-methyltetrahydrofolate; Inhibitor
Barbiturates are the first line drugs for treatment of epilepsy for adults and children in the developing world because of its low cost and proven effectiveness. Due to their adverse effects, no effective management of toxicity and difficulty in determining correct dosage, benzodiazepines are preferred over them. It is also used as a recreational drug, thereby contributing to cases of overdose [1]. My present study was to find the inhibitors of barbiturates by in-silico molecular docking. This approach can highlight only competitive inhibitors as molecules in-silico are rigid and do not produce conformational changes in structure by allosteric binding [2]. Gleobacter Ligand-Gated Ion Channel (GLIC) tertiary structure 5L47 [3] was downloaded from Protein Data Bank (Figure 1). FDAapproved drug structures were downloaded from Zinc15 database [4] consisting of around 450 compounds. Selenocyanobarbital was the desired ligand in GLIC structure (Figure 2). The target active site to which selenocyanobarbital binds was visualized using Drug Discover Studio [5]. After determining the amino acids of the target active site, the structure was cleaned of water molecules, ligands and other heterogenous atoms or molecules. Thiopental was chosen as the reference barbiturate molecule. Its three-dimensional structure was downloaded from Zinc15 database. PyRx software [6] was used for molecular docking because of its graphical user interface and inclusion of two important software, Autodock Vina [7] and Open Babel [8]. The operations were performed on a Windows 10 operating system utilizing 8 processors. All drug structures obtained from Zinc15 were processed in Open Babel to convert them to the minimum energy conformation. Partial charges and hydrogen were added and the structures were converted into Autodock ligands. The GLIC structure was then processed into Autodock molecule using PyRx. Autodock Vina wizard was used to dock thiopental to the active site determined before. The search space was adjusted in three-dimension to avoid undesirable binding. Exhaustiveness was kept at the maximum of 8. Of the 8 modes in which thiopental bonded, the first mode which had Root Mean Square Deviation (RMSD) of 0.00 was chosen as reference. The binding affinity of this mode E0 was chosen as the reference binding energy. In the same way, all the FDA-approved drug structures underwent molecular docking. Their binding affinity with RMSD equal to 0.00 was compiled. Binding affinity (k cal/mol) in negative signifies stronger binding. Hence, more the value in negative more is the binding affinity. Therefore top 35 ligands which had a more binding affinity than thiopental E0 are chosen for deep analysis. The ligands/drugs which are banned for use in India were removed from the list. Since ligands can interact with other amino acids in search space other than the target amino acids, these interactions with the receptor/GLIC were visualized using Discovery Studio. The number of desired interactions achieved was noted and then the results were sorted to provide the final candidates which interacted with at least 4 of the 6 amino acids. All the above steps taken improve the accuracy of the results which is desired. There were 6 active site amino acids determined from the interaction between Selenocyanobarbital and GLIC. These are listed as follows and represented in the diagram (Figure 3).
Figure 1: Gleobacter Ligand-gated Ion Channel (GLIC): 5L47 from Protein Data Bank. Different colours represent different chains.
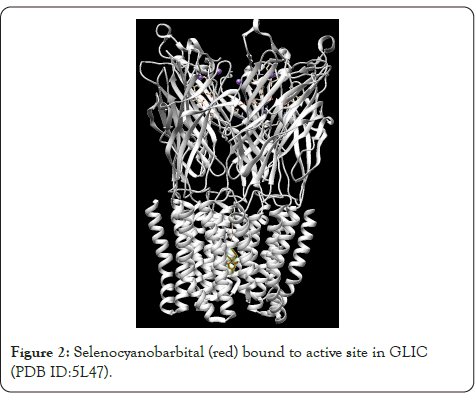
Figure 2: Selenocyanobarbital (red) bound to active site in GLIC (PDB ID:5L47).
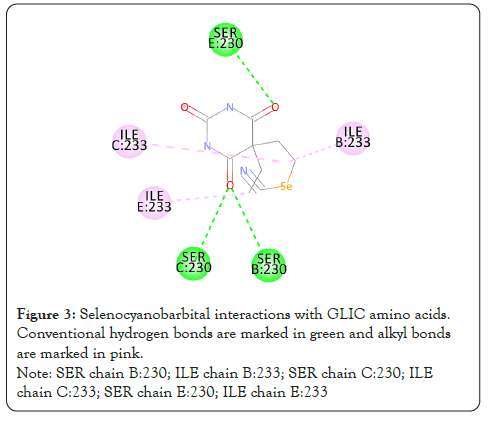
Figure 3: Selenocyanobarbital interactions with GLIC amino acids. Conventional hydrogen bonds are marked in green and alkyl bonds are marked in pink. Note: SER chain B:230; ILE chain B:233; SER chain C:230; ILE chain C:233; SER chain E:230; ILE chain E:233
ILE and SER are standard abbreviations for amino acids isoleucine and serine respectively. Numbers indicate their position in the chain. Active sites in GLIC are selected for docking in PyRx. Autodock Vina search space was restricted to the following coordinates taking care to include all active sites. The search space co-ordinates were as follows:
E0 binding affinity of thiopental was -6.1 kcal/mol. On the basis of the conditions set above, the final set of ligands obtained was as follows (Table 1).
| Ligand | Binding Affinity (k cal/mol) | Name (as per Zinc15 database) | Favourable interactions out of 6 |
|---|---|---|---|
| 5l47_clean_ZINC000001530948_uff_E=423.88 | -8.8 | Thalomid | 6 |
| 5l47_clean_ZINC000000004724_uff_E=549.93 | -7.6 | Trileptal | 4 |
| 5l47_clean_ZINC000000004785_uff_E=505.01 | -7.4 | Tegretol | 4 |
| 5l47_clean_ZINC000000136138_uff_E=288.75 | -7.4 | Oxybenzone | 5 |
| 5l47_clean_ZINC000002005305_uff_E=320.58 | -7.4 | 5-methyltetrahydrofolate | 4 |
| 5l47_clean_ZINC000000057512_uff_E=649.79 | -7.2 | Tolazamide | 6 |
| 5l47_clean_ZINC000001850377_uff_E=1617.03 | -7.2 | Nix | 4 |
| 5l47_clean_ZINC000002570817_uff_E=269.88 | -7.1 | Bromfenac | 5 |
| 5l47_clean_ZINC000000002191_uff_E=383.21 | -7 | Tolmetin | 4 |
| 5l47_clean_ZINC000000002279_uff_E=556.56 | -7 | Ketorolac | 4 |
| 5l47_clean_ZINC000000089763_uff_E=682.49 | -7 | Gantanol | 4 |
| 5l47_clean_ZINC000000518554_uff_E=227.11 | -7 | Arbutin | 5 |
| 5l47_clean_ZINC000000002216_uff_E=197.08 | -6.9 | Hand | 4 |
| 5l47_clean_ZINC000000120319_uff_E=493.79 | -6.9 | Sda | 5 |
| 5l47_clean_ZINC000000154964_uff_E=300.24 | -6.9 | Mepivacaine | 5 |
Table 1: Top 15 ligands arranged in increasing order of binding affinity along with the number of desired interactions greater than or equal to 4.
The interactions are represented in the 2-dimensional diagram below (Figures 4-18). The main objective of this study is to find an agent which can reverse the effect the barbiturate. Literature search for all the above compounds were done to find any documented relation between them and barbiturates. Thalomid which is the trade name for Thalidomide is used as a non-barbiturate sedative. It also enhances the effects of barbiturate [9] and hence is against the objective. Trileptal and tegretol are trade names for oxcarbazepine and carbamazepine respectively. They are used as seizure control drugs [10,11] and preferred over barbiturate but in no way can reverse the effect of barbiturate as they have the same clinical action. Tolazamide which is an oral anti-diabetic drug has been documented to interfere with barbiturate [12,13]. This is highly probable due to the fact that they both target similar active site found in this study. It can hence serve as competitive inhibitor of barbiturates. 5-methyltetrahydrofolate which is the active metabolite of folic acid has been experimented upon in the past and has shown inverse relation with phenytoin and barbiturate level in the body [14]. Patients receiving anti-convulsant therapy consisting of phenytoin and barbiturate had developed megaloblastic anaemia due to folate deficiency [15]. Gantanol which is trade name for sulfamethoxazole had only one study with relation to barbiturates [16]. However, that study could not be accessed. No relevant literature could be found for other compounds of this study. The final two candidates after this study are tolazamide and 5-methyltetrahydrofolate. These drugs can be used in future for clinical trials for management of barbiturate overdose.
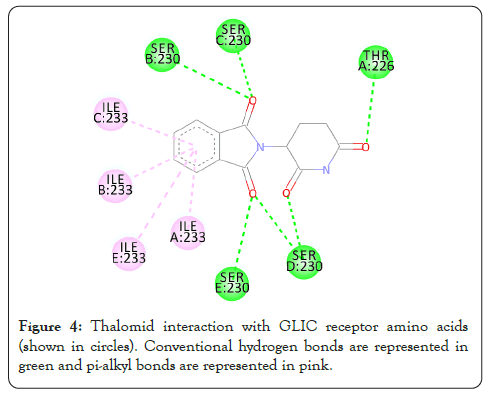
Figure 4: Thalomid interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
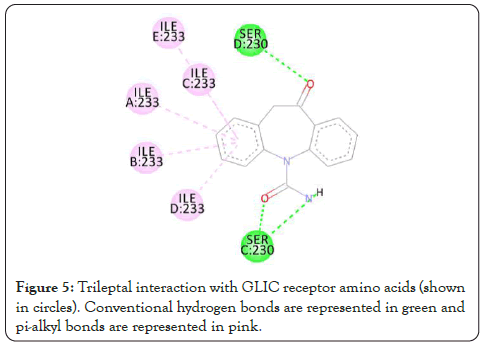
Figure 5: Trileptal interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
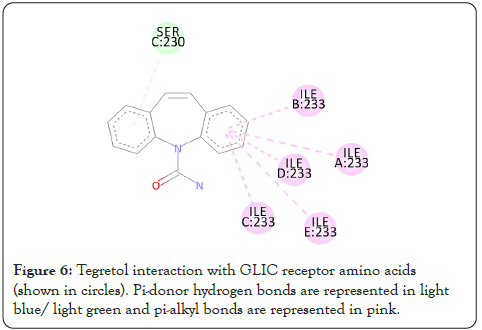
Figure 6: Tegretol interaction with GLIC receptor amino acids (shown in circles). Pi-donor hydrogen bonds are represented in light blue/ light green and pi-alkyl bonds are represented in pink.
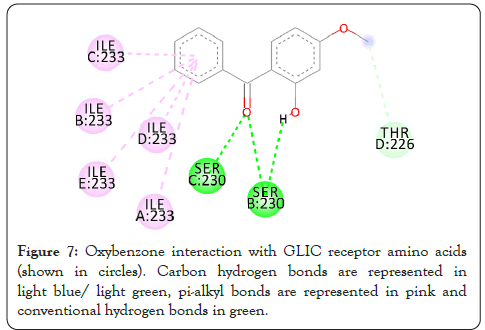
Figure 7: Oxybenzone interaction with GLIC receptor amino acids (shown in circles). Carbon hydrogen bonds are represented in light blue/ light green, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
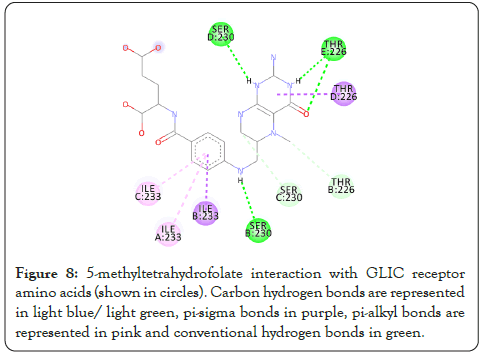
Figure 8: 5-methyltetrahydrofolate interaction with GLIC receptor amino acids (shown in circles). Carbon hydrogen bonds are represented in light blue/ light green, pi-sigma bonds in purple, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
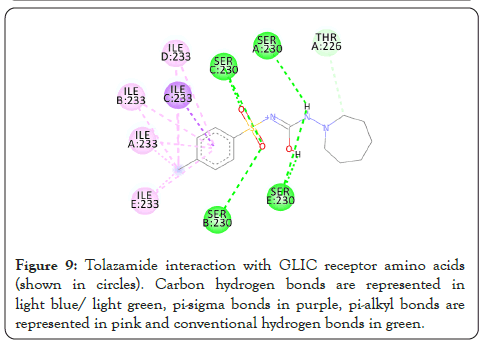
Figure 9: Tolazamide interaction with GLIC receptor amino acids (shown in circles). Carbon hydrogen bonds are represented in light blue/ light green, pi-sigma bonds in purple, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
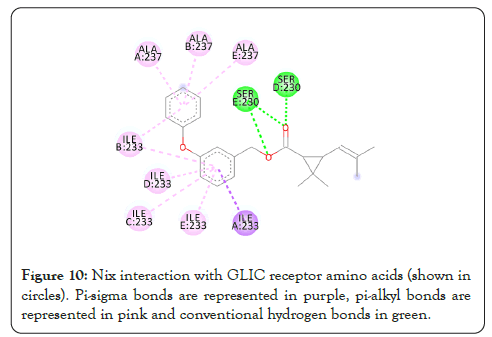
Figure 10: Nix interaction with GLIC receptor amino acids (shown in circles). Pi-sigma bonds are represented in purple, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
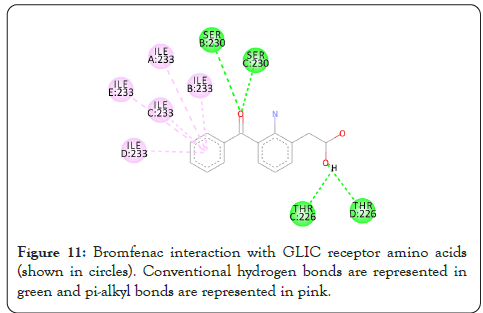
Figure 11: Bromfenac interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
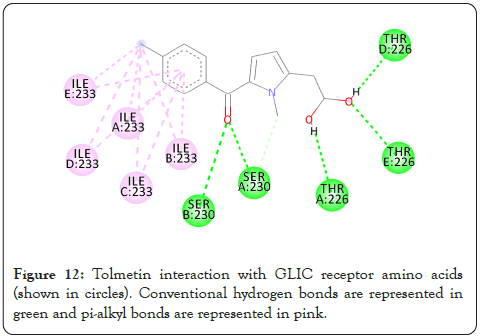
Figure 12: Tolmetin interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
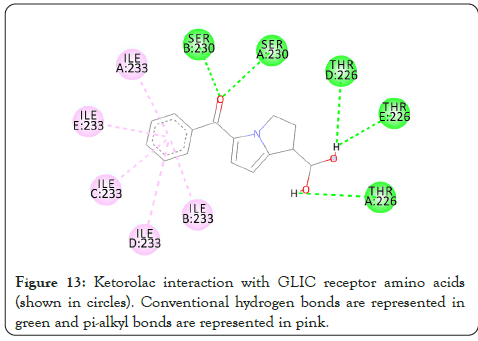
Figure 13: Ketorolac interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
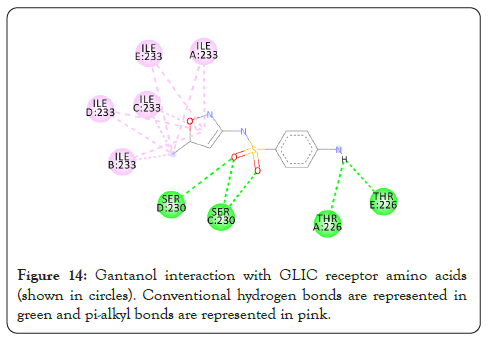
Figure 14: Gantanol interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
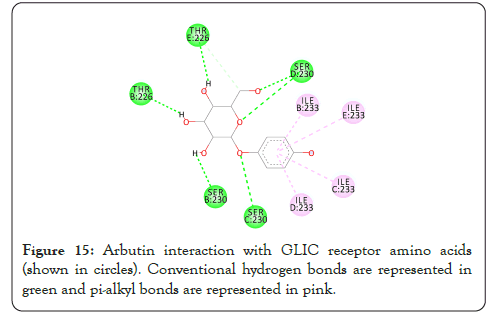
Figure 15: Arbutin interaction with GLIC receptor amino acids (shown in circles). Conventional hydrogen bonds are represented in green and pi-alkyl bonds are represented in pink.
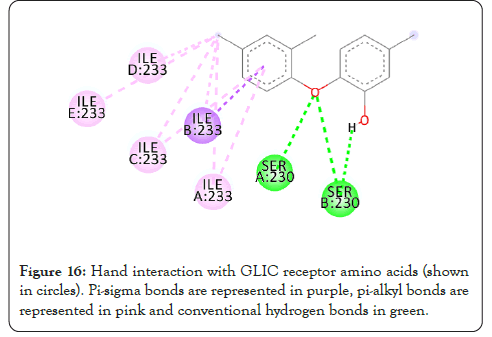
Figure 16: Hand interaction with GLIC receptor amino acids (shown in circles). Pi-sigma bonds are represented in purple, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
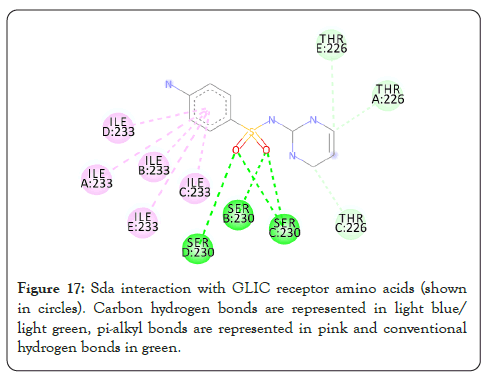
Figure 17: Sda interaction with GLIC receptor amino acids (shown in circles). Carbon hydrogen bonds are represented in light blue/ light green, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
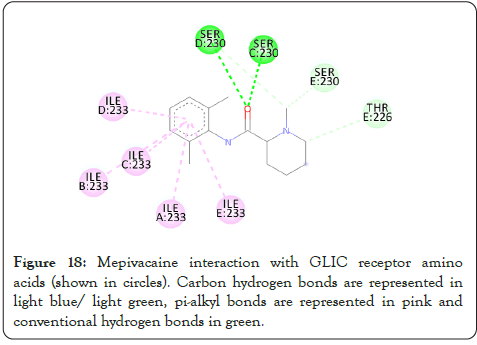
Figure 18: Mepivacaine interaction with GLIC receptor amino acids (shown in circles). Carbon hydrogen bonds are represented in light blue/ light green, pi-alkyl bonds are represented in pink and conventional hydrogen bonds in green.
The above study has brought out probable treatment of barbiturate overdose as tolazamide and 5-methyltetrahydrofolate. Antidotes for drugs need to exist as long as they are used for public. Clinical trials are however necessary to validate these results. Non-competitive inhibitors have to be found by in-vitro trials as it is the most important limitation of in-silico approach.
Nil.
Not required.
None.
[Crossref]
Citation: Sharp K (2022) Folate and Tolazamide: Potential Competitive Inhibitors of Barbiturate by In-Silico Molecular Docking. Enz Eng. 11:187.
Received: 01-Jun-2022, Manuscript No. EEG-22-17696; Editor assigned: 03-Jun-2022, Pre QC No. EEG-22-17696 (PQ); Reviewed: 21-Jun-2022, QC No. EEG-22-17696; Revised: 27-Jun-2022, Manuscript No. EEG-22-17696 (R); Published: 04-Jul-2022 , DOI: 10.35841/2329-6674.22.11.187
Copyright: © 2022 Sharp K. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.