Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2022)Volume 12, Issue 6
Background: Ribosomal DNA (rDNA), consisting of the 18S, 5.8S, 25S and 5S rRNA genes and noncoding functional elements in eukaryotes, is a region in the genome with very high activity with respect to transcription and genomic rearrangement. Due to its highly repetitive and complex structure, detailed sequence analyses are challenging and often neglected. In this study, a systematic survey of rDNA organization across the fungal kingdom was performed.
Methods: We analysed all fungal genome assemblies available at NCBI, in total 6779, by bioinformatic methods. For this, we developed a batch of Python scripts to process the data. For de-novo assembling of NGS data, we wrote the Ansi C program ‘YNGS’.
Results: Only 15% of the fungal genome assemblies at NCBI possessed the sequence of an entire rDNA unit. Most assemblies of the rDNA were incomplete and were interrupted in the internal spacer regions. Next, we analysed the rDNA organisation. Basidiomycota, with the exception of the Ustilaginomycetes, had 5S gene inserted in the rDNA units as well Blastocladiomycota, Chytridiomycota and Mucoromycota. Ascomycota, with the exception of Saccharomycetes, lacked 5S insertions in the rDNA units. Here, the 5S genes were scattered in the genome and tandem arrays of 5S genes were very uncommon. With the YNGS software, we assembeled 17 entier rDNA unities and calculated an amount of 25 to 161 rDNA repeated units per genome.
Conclusion: In contrast to the rDNA gene region, the internal spacer regions are still challenging to analyse, even in the NGS era. Transcriptional regulation, replication machinary and genome rearrengment elements are meeting together. Sophisticate regulations are necessary for a smooth flow.
Genome; rDNA; 5S insertion; Mycology; Bioinformatics
Ribosomal RNAs (rRNAs) are essential macro-molecules and are present in all living cells, In Bacteria, Archaea and Eukaryota. Together with protein, they are forming together the ribosomes, the machinery translating mRNA into protein. For the correct translation, a matching tRNA docks to the mRNA template at the small sub-unit (SSU), whereas the large sub-unit (LSU) facilitates the process of protein synthesis. In Eukaryota, the SSU contains a single RNA molecule, the 18S, whereas the LSU has three different kinds of rRNA molecules, 5S, 5.8S and 25S (Table 1). For historical reasons, the different rRNAs are named according to their sedimentation coefficients measured in Svedberg units (S). Unfortunately, the use of this unit leads to inconsistency in the nomenclature of the rRNA molecules, since the sedimentation coefficient of homologous rRNA differs among taxa. Following, we are using the nomenclature of Saccharomyces cerevisiae.
| Fungi*I | Human*II | Bacteria*III | Subunits | |
|---|---|---|---|---|
| 80S | 80S | 70S | Ribosome | |
| 60S | 60S | 50S | large subunit (LSU) of the ribosome | |
| 40S | 40S | 30S | small subunit (SSU) of the ribosome | |
| 35S | 45S | 30S | pre-rRNA molecule; before splicing | |
| 25S | 28S | 23S | rRNA of LSU; transcribed by the RNA polymerase I | |
| 18S | 18S | 16S | rRNA of the SSU; transcribed by the RNA polymerase I | |
| 5.8S | 5.8S | - | rRNA of LSU; transcribed by the RNA polymerase I | |
| 5S | 5S | 5S | rRNA of LSU, RNA polymerase III | |
Note: *I based on Saccharomyces cerevisiae, *II based on Homosapiens, *III based on Escherichia coli.
Table 1: Common rDNA nomenclature in fungi, animals and bacteria.
rDNA organization in the eukaryotic genome
In eukaryotic organisms, ribosomal genes are present in multiple repeat units to satisfy the high demand for ribosomes. They are organized in one to a few arrays (Figure 1A). In plants and animals, 5S rDNA genes are commonly separated from main rDNA arrays in Figure 1B, but are commonly organized in separate arrays of tandem repeats (Potapova and Gerton) [1].
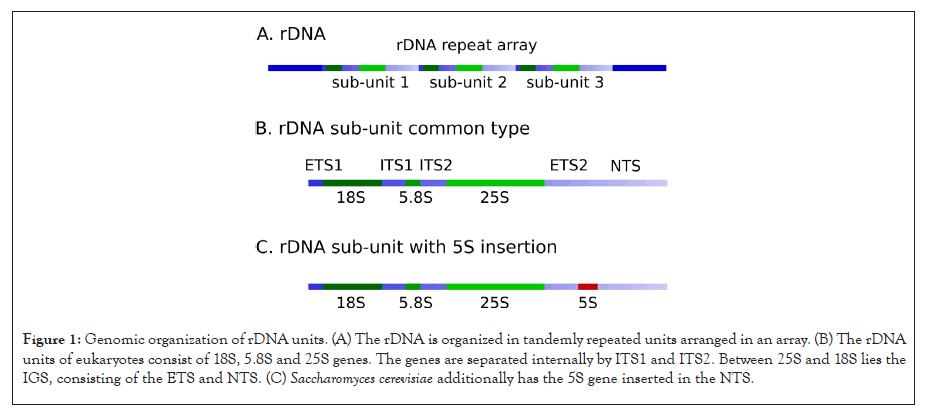
Figure 1: Genomic organization of rDNA units. (A) The rDNA is organized in tandemly repeated units arranged in an array. (B) The rDNA units of eukaryotes consist of 18S, 5.8S and 25S genes. The genes are separated internally by ITS1 and ITS2. Between 25S and 18S lies the IGS, consisting of the ETS and NTS. (C) Saccharomyces cerevisiae additionally has the 5S gene inserted in the NTS.
rDNA copy number varies between 39 and 19 300 in animals and from 150 to 26 048 in plants, whereby the copy number and genome size of the organism are strongly correlated (Prokopowich et al.) [2]. However, large variations in gene numbers exist not only among closely related species but also among different strains of the same taxon (Long and Dawid et al.) [3].
Challenges in studying rDNA organization
Most bioinformatic studies focus on low-copy DNA regions. Analyses of repeated sequences are often neglected. In the final assemblies, the repeated sequences are omitted, are collapsed to a few repeated units or cause interruptions in the contigs (Biscotti et al.) [4]. From a computational perspective, highly repeated sequences can create ambiguities in alignments and can subsequently create errors in genome assemblies (Treangen and Salzberg) [5]. De novo assembly software frequently fails to resolve rDNA sequences. Even when combining NGS (Illumina and 454) data, Sanger sequences and BLAST searches against the National Center for Biotechnology Information (NCBI) database, only one of five programs worked efficiently (Agrawal and Ganley) [6].
rDNA within fungal model organisms
The genomes of the four most intensively studied fungal model species were resolved at the chromosome level. To accomplish this, the assemblies were based on various sequencing data, next- generation sequencing (NGS), bacterial artificial chromosome (BAC), Fosmid, Cosmid and Sanger sequencing data.
Saccharomyces cerevisiae: Each rDNA unit, with a length of 9.1 kB, consists of one 5S, 5.8S, 15S and 25S gene (Figure 1C). The genes are separated by three kinds of spacers: nontranscribed spacers (NTS1 and NTS2), external transcribed spacers (ETS1 and 3' ETS2) and internal transcribed spacers (ITS1 and ITS2). The rDNA is organized into 100-200 tandem repeats on chromosome XII. In the reference genome assembly (R64-2- 1), only two units of the array are displayed (www.yeastgenome.org, Christie et al.) [7].
Candida albicans: The arrangement of the rDNA genes in C. albicans is similar to that in S. cerevisiae but includes an additional low-complexity region of approximately 2 kb. Each of the approximately 55 units, estimated for a haploid set of chromosomes, is 12.8 kB long (Jones et al.) [8]. In the C. albicans ‘SC5314’ genome assembly, only one unit is displayed (www.candidagenome.org, Skrzypek et al.) [9].
Schizosaccharomyces pombe: In S. pombe, the rDNA is arranged in two tandem arrays, estimated to be 1225 kb and 240 kb in size. In the genome assembly ASM294v2 of strain 972h-, the two tandem arrays flank both ends of chromosome III and are only partially displayed. Distal flanking regions are missing in the assembly (www.pombase.org, Lock et al.) [10]. The 5S genes are, in contrast to those in S. cerevisiae and C. albicans, dispersed within the genome (Mao et al.) [11]. The six copies of 5S rDNA (SPRRNA.03, 04, 05, 06, 07 and 10) are not linked to each other.
Aspergillus nidulans: The rDNA units of A. nidulans are relatively short, with a size of 7.7 kB. The intergenic spacer (IGS), consisting of the NTS and ETS, is just 1.7 kB long (Lockington et al.) [12]. In the genome assembly at Aspergillus-database, four of the rDNA units are displayed, whereas the 5S genes are not mapped (www.aspergillusgenome.org, Cerqueira et al.) [13]. In A. nidulans, the 5S rDNA genes are dispersed throughout the genome and are not organized in arrays. Six 5S rDNA fragments are present, four have complete 5S rRNA sequences and two are pseudogenes (Bartnik et al.) [14].
All calculations were performed with a ThinkPad P52 equipped with an i7 processor and 64 GB RAM. The operating system was a Debian Release 10 Linux operating system with Python 3.7 and GCC version 8.3.
Fungal assemblies, rDNA annotation and analysis
Four datasets of fungal assemblies were downloaded from the NCBI (Table 2). To analyse the DNA sequences, we wrote six Python scripts in Table 3 and run the scripts successively (Figure 2A). First, we created a list of all genome assemblies, whereby duplicate entries were removed, by successively running Python scripts #1 and #2. In the next step, we annotated the rDNA genes with RNAmmer version 1.2 software (Lagesen et al.) [15]. For automatic analysis of the datasets, Python script #3 unzipped each assembly and consecutively called RNAmmer software. Next, we sorted the rDNA annotations with Python script #4. To correlate the rDNA organization in the genome with phylogenetic information, we downloaded lineage information from the NCBI Taxonomy Database for each assembly and created a list with information about the kingdom, phylum, class, order, family and genus (Python script #5). Python script #6 analysed the data and combined the results with phylogenetic information.
| Database name | Release Type | Filter | Entries |
|---|---|---|---|
| GenBank_total | GenBank | fungi, latest | 6453 |
| GenBank_genome | GenBank | fungi, latest and complete genome | 72 |
| RefSeq_total | RefSeq | fungi, latest | 326 |
| RefSeq_genome | RefSeq | fungi, latest and complete genome | 11 |
Table 2: Databases of fungal assemblies downloaded from the NCBI assembly database.
| Python | Database |
|---|---|
| #1 | Create a list of assembly name, fasta file comment of each contig and contig length of the gzipped fasta files |
| #2 | Computes as list of assembly name, species name and database |
| #3 | Calls the software RNAmmer on a batch of gzipped fasta files |
| #4 | Python script for summarizing and preprocessing RNAmmer results |
| #5 | Python script for getting the lineage, phylogeny, for all genome assemblies |
| #6 | Create a list of the maximal 35S and 5S rDNA repeat amount for each assembly and add lineage information |
#The rDNA genes that analysing in NCBI database.
Table 3: Phyton scripts for annotating and analysing rDNA genes in assemblies downloaded from the NCBI database.
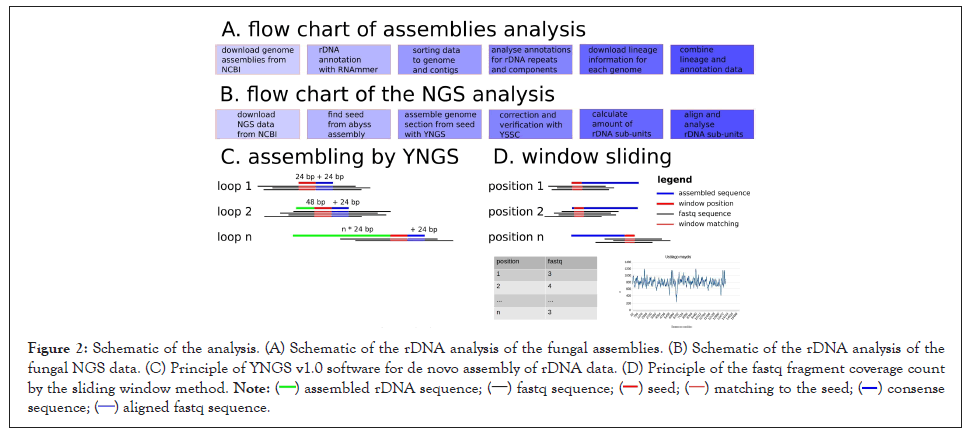
Figure 2: Schematic of the analysis. (A) Schematic of the rDNA analysis of the fungal assemblies. (B) Schematic of the rDNA analysis of the fungal NGS data. (C) Principle of YNGS v1.0 software for de novo assembly of rDNA data. (D) Principle of the fastq fragment coverage count by the sliding window method.
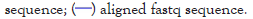
rDNA sequences assembled from fastq files
For de novo assembly of rDNA repeat units, we downloaded NGS data in Table 4 from the Sequence Read Archive (SRA) of NCBI. For the de novo assembly of rDNA sequences, we wrote YNGS v1.0 software and released it as open source software at Bioinformatics.org. YNGS software assembles DNA sequences from a 50 bp seed by elongating the sequence with 24 bp per loop (Figure 2C). In cases of interruption of the elongation, due to parts of the sequence that are difficult to align, the fastq forwards and paired-end sequences were displayed in a separate window and the string was elongated manually. Afterwards, the quality of the de novo assemblies were verified by counting the fastq fragments matching each position of the assembly (Figure 2D) with the sliding window approach.
| Taxon | sra file downloaded from NCBI |
|---|---|
| Phaeotremella foliacea | DRR032527.sra |
| Cerinomyces ceraceus | SRR4151924.sra |
| Boletus edulis | SRR5764827.sra |
| Moesziomyces sp. | SRR6053896.sra |
| Ustilago maydis | DRR032503.sra |
| Rhodotorula mucilaginosa | SRR11859272.sra |
| Verticillium dahliae | SRR12769281.sra |
| Sordaria macrospora | SRR5749461.sra |
| Morchella importuna | SRR4136319.sra |
| Candida auris | SRR12784121.sra |
| Saccharomyces cerevisiae | SRR4244871.sra |
| Candida parapsilosis | SRR8984820.sra |
| Schizosaccharomyces pombe | SRR11003486.sra |
| Taphrina betulina | SRR10079519.sra |
| Paraphysoderma sedebokerense | SRR6447920.sra |
| Allomyces javanicus | SRR11025671.sra |
| Geranomyces variabilis | SRR12278674.sra |
| Zygnema circumcarinatum | SRR8552832.sra |
| Chlorella vulgaris | SRR8083359.sra |
| Boletus edulis | SRR5764827.sra |
| Moesziomyces sp. | SRR6053896.sra |
| Ustilago maydis | DRR032503.sra |
| Rhodotorula mucilaginosa | SRR11859272.sra |
| Verticilliumdahliae | SRR12769281.sra |
| Sordaria macrospora | SRR5749461.sra |
| Morchella importuna | SRR4136319.sra |
| Candida auris | SRR12784121.sra |
| Saccharomyces cerevisiae | SRR4244871.sra |
| Candida parapsilosis | SRR8984820.sra |
| Schizosaccharomyces pombe | SRR11003486.sra |
| Taphrina betulina | SRR10079519.sra |
| Paraphysoderma sedebokerense | SRR6447920.sra |
| Allomyces javanicus | SRR11025671.sra |
| Geranomyces variabilis | SRR12278674.sra |
| Zygnema circumcarinatum | SRR8552832.sra |
| Chlorella vulgaris | SRR8083359.sra |
Table 4: Fastq files downloaded from the NCBI SRA database for de novo assembling.
The rDNA sequences were aligned with Clustalx v2.1 software. For the annotation of the rDNA genes, data for the model organism S. cerevisiae were used as a template. The phylogenetic dendrogram was created with the sequence from the 35S gene with Phylip v3.697 software. The alignment was bootstrapped with 100 replications. For the calculation of repeats per genome, we identified the fastq fragments matching the rDNA sequence. For normalization, we used the four genes ACT1, TRP3, TUB4 and URA3. We calculated the number of fastq fragments matching the four sequences with the sliding window application in YNGS software. Finally, we divided the average amount of rDNA matching fastq fragments with the amount of the normalizing genes.
To examine the genomic organization of the rDNA in fungal species, we analysed two different data sources. First, we analysed fungal genomic assemblies, which were available in the NCBI database. The high number of available datasets allowed a systematic study of rDNA organization. Second, we focused on a few manually selected species and assembled the rDNA units.
Fungal assemblies
A total of 6779 fungal assemblies were available in the NCBI databank. Of these, 6453 were in the GenBank dataset, and 326 were in the RefSeq dataset. In the GenBank dataset, 72 were assembled to genome quality, and in the RefSeq dataset, 11. With RNAmmer software, 5S genes were annotated in 6050 of the total 6779 assemblies, 18S genes in 4776 of the assemblies and 25S genes in 4556 of the assemblies (Table 5). In 1032 of the assemblies, at least one entire repeat unit was present. In 603 assemblies, at least three repeating units were present (Figure 3). Among all assemblies, less than 9% possessed at least three complete units, among those in GenBank, less than 9%, and among those in RefSeq, less than 14%. In the assemblies with genome quality, 61% of the GenBank and 36% of the RefSeq assemblies had at least three entire units in a row.
Figure 3: Annotation of rDNA genes in the fungal assemblies. The rDNA genes of 6779 assemblies were annotated. A total of 6050 assemblies had 5S genes, 4776 had 18S, and 4556 had 25S annotations. In 1409 of the assemblies, at least one complete rDNA unit in an array was annotated, and in 247 assemblies, at least three units in an array were annotated.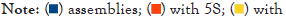
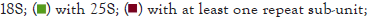

| Assemblies | total | GenBank | GenBank genome | RefSeq | RefSeq genome |
|---|---|---|---|---|---|
| entries | 6779 | 6453 | 72 | 326 | 11 |
| 5S*I | 6050 | 5747 | 66 | 303 | 11 |
| 18S*II | 4776 | 4574 | 67 | 202 | 11 |
| 25S*III | 4556 | 4359 | 66 | 197 | 11 |
| at least one repeat*IV | 1032 | 946 | 56 | 86 | 7 |
| at least three repeats*V | 603 | 559 | 44 | 44 | 4 |
Note:*I amount of assemblies with at least one 5S rDNA gene annotation.
*II amount of assemblies with at least one 18S rDNA gene annotation.
*III amount of assemblies with at least one 25S rDNA gene annotation.
*IV amount of assemblies with at least one entire repeat unit of rDNA.
*V amount of assemblies with at least three repeat units of rDNA in a continuous row.
Table 5: Annotation of rDNA genes and repeat length of the genome assemblies.
Organization of the rDNA array in the genome
Based on the annotation of the rDNA genes, we identified the rDNA arrays having 5S insertions in the intergenic spacer region. Therefore, we selected the largest rDNA array from each of the 603 assemblies. The 5S insertions were present in 298 of the arrays and were absent in 305.
To correlate the presence of 5S genes with phylogenetic data, we downloaded the lineage information for each taxon from the NCBI database. Most of the Basidiomycota had 5S insertions in the rDNA repeat units, whereas most of the Ascomycota lacked 5S insertions. For the phyla Blastocladiomycota, Chytridiomycota and Mucoromycota, five assemblies could be evaluated, and in all cases, 5S insertions were present in the rDNA arrays (Figure 4). We sub-classified the data to the class level. In the phylum Ascomycota, nearly all assemblies, with the exception of Saccharomycetes, lacked 5S insertions in the rDNA repeats. In more than 98% of the analysed Ascomycota in the classes Dothideomycetes, Eurotiomycetes, Leotiomycetes, Orbiliomycetes, Pezizomycetes, Schizosaccharomycetes and Sordariomycetes, 5S insertions were absent (Table 6). In contrast to the other classes of Ascomycota, most taxa of the Saccharomycetes had 5S insertions. In 46 taxa, 5S insertions were present (Table 7). The 11 aberrant assemblies belonged the genera Hyphopichia, Komagataella, Metschnikowia, Sugiyamaella and Yarrowia in Table 6, accounting for ~6% of the total Saccharomycetes assemblies.
Figure 4: Phylogenetic groups with 5S insertions in the rDNA arrays. Basidiomycota, with the exception of the Ustilaginomycetes, have 5S insertions in the rDNA arrays. Ascomycota, with the exception of the Saccharomycetes, lack 5S insertions in the rDNA arrays.
| Dothideomycetes |
| Acidomyces richmondensis, Alternaria alternata, Alternaria brassicae, Alternaria brassicicola, Alternaria gaisen, Alternaria solani, Amniculicola lignicola, Ascochyta lentis, Ascochyta rabiei, Ascochyta versabilis, Bipolaris sorokiniana, Botryosphaeria dothidea, Cucurbitaria berberidis, Didymella segeticola, Dothidotthia symphoricarpi, Hortaea werneckii, Karstenula rhodostoma, Leptosphaeria biglobosa, Leptosphaeria maculans, Lizonia empirigonia, Macrophomina phaseolina, Paraphaeosphaeria minitans, Parastagonospora nodorum, Peltaster fructicola, Phoma sp., Pseudopyrenochaeta lycopersici, Pyrenophora teres, Pyrenophora tritici- repentis, Venturia inaequalis, Zymoseptoria tritici |
| Eurotiomycetes |
| Ajellomyces capsulatus, Arthroderma uncinatum, Aspergillus awamori, Aspergillus campestris, Aspergillus cejpii, Aspergillus cristatus, Aspergillus flavus, Aspergillus fumigatus, Aspergillus lentulus, Aspergillus nidulans, Aspergillus niger, Aspergillus ochraceus, Aspergillus oryzae, Aspergillus persii, Aspergillus steynii, Aspergillus terreus, Aspergillus tubingensis, Blastomyces dermatitidis, Blastomyces gilchristii, Coccidioides immitis, Coccidioides posadasii, Emergomyces orientalis, Exophiala lecanii-corni, Exophiala spinifera, Microsporum canis, Monascus purpureus, Paracoccidioides brasiliensis, Paracoccidioides lutzii, Paracoccidioides sp., Penicillium capsulatum, Penicillium expansum, Penicillium oxalicum, Penicillium solitum, Penicillium sp., Talaromyces funiculosus, Talaromyces marneffei, Talaromyces pinophilus, Uncinocarpus reesii |
| Leotiomycetes |
| Articulospora tetracladia, Blumeria graminis, Ciboria shiraiana, Helotiales sp., Podosphaera xanthii, Pseudogymnoascus destructans, Sclerotinia sclerotiorum |
| Orbiliomycetes |
| Arthrobotrys flagrans, Arthrobotrys oligospora |
| Pezizomycetes |
| Morchella sextelata, Tuber indicum |
| Schizosaccharomycetes |
| Schizosaccharomyces japonicus, Schizosaccharomyces octosporus, Schizosaccharomyces pombe |
| Sordariomycetes |
| Amesia nigricolor, Amphirosellinia nigrospora, Aquanectria penicillioides, Beauveria bassiana, Ceratocystis albifundus, Cladobotryum protrusum, Colletotrichum acutatum, Colletotrichum destructivum, Colletotrichum fructicola, Colletotrichum gloeosporioides, Colletotrichum higginsianum, Colletotrichum siamense, Colletotrichum sp., Colletotrichum sublineola, Colletotrichum tofieldiae, Colletotrichum trifolii, Colletotrichum viniferum, Coniochaeta sp., Cordyceps militaris, Cryphonectria parasitica, Daldinia childiae, Daldinia concentrica, Dicyma pulvinata, Entonaema liquescens, Epichloe festucae, Esteya vermicola, Fragosphaeria purpurea, Fusarium culmorum, Fusarium graminearum, Fusarium oxysporum, Fusarium venenatum, Fusarium verticillioides, Gaeumannomyces sp., Grosmannia penicillata, Hypomontagnella monticulosa, Hypomyces perniciosus, Hypomyces rosellus, Hypoxylon fragiforme, Hypoxylon lienhwacheense, Hypoxylon pulicicidum, Magnaporthales sp., Magnaporthe oryzae, Neurospora crassa, Neurospora sp., Ophiocordyceps camponoti- floridani, Ophiocordyceps sinensis, Pyrenopolyporus hunteri, Pyricularia oryzae, Raffaelea albimanens, Raffaelea quercivora, Reticulascus tulasneorum, Sarocladium brachiariae, Tolypocladium inflatum, Tolypocladium sp., Trichoderma harzianum, Trichoderma reesei, Trichoderma sp., Trichoderma virens, Verticillium albo-atrum, Verticillium alfalfae, Verticillium dahliae, Xylaria grammica, Xylaria hypoxylon, Xylaria sp. |
| Saccharomycetes |
| Hyphopichia burtonii, H. pseudoburtonii, Komagataella pastoris, K. phaffii, M. citriensis, M.reukaufii, M. reukaufii, M. sp., Sugiyamaella lignohabitans, Yarrowia lipolytica |
Table 6: Ascomycota taxa without 5S insertions in the rDNA arrays.
| Saccharomycetes |
|---|
| Ashbya gossypii, Brettanomyces custersianus, Brettanomyces nanus, Candida albicans, Candida auris, Candida duobushaemulonis, Candida glabrata, Candida metapsilosis, Candida parapsilosis, Candida sake, Candida sojae, Candida tropicalis, Candida vartiovaarae, Candida viswanathii, Clavispora lusitaniae, Eremothecium gossypii, Eremothecium sinecaudum, Hanseniaspora uvarum, Kazachstania servazzii, Kluyveromyces lactis, Kluyveromyces marxianus, Kodamaea ohmeri, Lodderomyces elongisporus, Magnusiomyces capitatus, Magnusiomyces ingens, Meyerozyma guilliermondii, Millerozyma farinosa, Ogataea polymorpha, Pichia kluyveri, Pichia kudriavzevii, Pichia manshurica, Saccharomyces cerevisiae, Saccharomyces eubayanus, Saccharomyces paradoxus, Saccharomyces pastorianus, Saccharomyces sp., Saccharomycopsis fibuligera, Saccharomycopsis malanga, Saprochaete fungicola, Saprochaete ingens, Saprochaete suaveolens, Scheffersomyces stipitis, Torulaspora delbrueckii, Zygosaccharomyces mellis, Zygosaccharomyces rouxii, Zygosaccharomyces sapae |
Table 7: Ascomycota with 5S insertions in the rDNA arrays.
In Basidiomycota, 5S insertions in the rDNA arrays were very common Table 8, in Agaricomycetes, Malasseziomycetes, Microbotryomycetes, Moniliellomycetes, Pucciniomycetes and Tremellomycetes. In contrast, in the class Ustilaginomycetes, all 7 assemblies lacked 5S insertions in the rDNA arrays (Table 9). In the Mucoromycota in Table 10, Blastocladiomycota Table 11 and Chytridiomycota in Table 12, 5S insertions were present.
| Agaricomycetes |
| Agaricus bisporus, Agrocybe pediades, Amylostereum areolatum, Armillaria cepistipes, Armillaria gallica, Auricularia cornea, Auricularia heimuer, Coriolopsis trogii, Flammulina velutipes, Floccularia luteovirens, Ganoderma australe, Grammothele lineata, Hericium coralloides, Irpex lacteus, Lentinula edodes, Macrocybe gigantea, Phellinus noxius, Pleurotus eryngii, Pleurotus tuoliensis, Polyporus brumalis, Sanghuangporus sanghuang, Sarcomyxa edulis, Serpula lacrymans, Trametes hirsuta |
| Malasseziomycetes |
| Malassezia furfur, Malassezia globosa, Malassezia restricta, Malassezia sympodialis |
| Microbotryomycetes |
| Microbotryum intermedium, Microbotryum lychnidis-dioicae, Microbotryum saponariae, Microbotryum silenes- acaulis, Microbotryum violaceum, Rhodosporidium toruloides, Rhodotorula graminis, Rhodotorula kratochvilovae, Rhodotorula toruloides |
| Moniliellomycetes |
| Moniliella sp. |
| Pucciniomycetes |
| Hemileia vastatrix, Melampsora larici-populina, Puccinia graminis, Puccinia hordei, Austropuccinia psidii |
| Tremellomycetes |
| Apiotrichum porosum, Cryptococcus gattii, Cryptococcus neoformans, Cryptococcus sp., Saitozyma podzolica |
Table 8: Basidiomycota with 5S insertions in the rDNA arrays.
| Ustilaginomycetes |
|---|
| Anthracocystis flocculosa, Moesziomyces antarcticus, Sporisorium scitamineum, Thecaphora thlaspeos, Ustilago bromivora, Ustilago trichophora |
Table 9: Basidiomycota without 5S insertions in the rDNA arrays.
| Mucoromycetes |
|---|
| Absidia repens, Mucor lusitanicus |
Table 10: Mucoromycota with 5S insertions in the rDNA arrays.
| Blastocladiomycetes |
|---|
| Allomyces macrogynus, Catenaria anguillulae |
Table 11: Blastocladiomycota with 5S insertions in the rDNA arrays.
| Chytridiomycetes |
|---|
| Rhizoclosmatium globosum |
Table 12: Chytridiomycota with 5S insertions in the rDNA arrays.
Organization of 5S rDNA
In plants and animals, which commonly lack 5S insertions in rDNA arrays, 5S genes are frequently clustered in the genome and arranged in separated 5S gene arrays. To explore the distribution and size of 5S genes in the 603 fungal assemblies, we calculated the number of neighboring 5S genes (Table 13). In nearly all of the fungal assemblies, 5S gene arrays were not present. Most commonly, the 5S genes were single, separated and scattered in the genome (~79%). Only 8 assemblies (belonging to only four species) had arrays exceeding 5S gene repeats (Table 14). The largest 5S gene array was detected in assemblies of the powdery mildew Blumeria graminis, with 20 tandemly repeated 5S genes. Anthracocystis flocculosa, a member of Ustilaginales, had 18 repeats, giving it the second largest 5S gene array. The 5S gene arrays in S. cerevisiae were striking. The dataset contained 124 S. cerevisiae assemblies and nine of them possessed neighbouring 5S genes. Whereas extended 5S gene rDNA arrays were rare, shorter arrays of 2 or 3 repeated 5S genes were more frequent. In some species, we observed an enrichment of two neighbouring 5S genes. In three fungal species, Candida sake, Coccidioides immitis and Trichoderma virens, more than 30% of the 5S genes were arranged in arrays of two neighbouring 5S genes, without the presence of larger arrays.
| Amount neighboring 5S genes | Amount assemblies |
|---|---|
| 20 | 1 |
| 18 | 1 |
| 15 | 1 |
| 9 | 2 |
| 7 | 1 |
| 6 | 2 |
| 5 | 2 |
| 4 | 5 |
| 3 | 20 |
| 2 | 88 |
| 1 | 477 |
| 0 | 3 |
Table 13: 5S gene arrays in the fungal assemblies
| Species | Phylum | Order | Assembly | size of 5S gene array |
|---|---|---|---|---|
| Blumeria graminis | Ascomycota | Erysiphales | GCA_900237765 | 20 |
| Anthracocystis flocculosa | Basidiomycota | Ustilaginales | GCA_900322975 | 18 |
| Blumeria graminis | Ascomycota | Erysiphales | GCA_900519115 | 15 |
| Saccharomyces cerevisiae | Ascomycota | Saccharomycetales | GCA_001305865 | 9 |
| Blumeria graminis | Ascomycota | Erysiphales | GCA_900239735 | 9 |
| Saccharomyces cerevisiae | Ascomycota | Saccharomycetales | GCA_000773925 | 7 |
| Morchella sextelata | Ascomycota | Pezizales | GCA_009741755 | 6 |
| Saccharomyces cerevisiae | Ascomycota | Saccharomycetales | GCA_902192305 | 6 |
Table 14: Fungal taxa with extended 5S gene arrays.
rDNA sequences assembled from NGS fastq files
We assembled the complete rDNA units of seventeen fungal species from NGS sequencing data (Table 15). To root the phylogenetic dendrogram in Figure 5, we additionally assembled the rDNA units of two algal species. The alga Zygnema circumcarinatum belongs to the Zygnematophyceae, whereas Chlorella vulgaris belongs to the Trebouxiophyceae.
| Taxon | Phylum | Subphylum | Insertion | Repeats |
|---|---|---|---|---|
| Phaeotremella foliacea | Basidiomycota | Agaricomycotina | forw | 60 |
| Cerinomyces ceraceus | Basidiomycota | Agaricomycotina | rev | 93 |
| Boletus edulis | Basidiomycota | Agaricomycotina | forw | 64 |
| Moesziomyces sp. | Basidiomycota | Ustilaginomycotina | without | 51 |
| Ustilago maydis | Basidiomycota | Ustilaginomycotina | without | 68 |
| Rhodotorula mucilaginosa | Basidiomycota | Pucciniomycotina | forw | 55 |
| Verticilliumdahliae | Ascomycota | Pezizomycotina | without | 74 |
| Sordaria macrospora | Ascomycota | Pezizomycotina | without | 130 |
| Morchella importuna | Ascomycota | Pezizomycotina | without | 117 |
| Candida auris | Ascomycota | Saccharomycotina | rev | 25 |
| Saccharomyces cerevisiae | Ascomycota | Saccharomycotina | rev | 147 |
| Candida parapsilosis | Ascomycota | Saccharomycotina | rev | 43 |
| Schizosaccharomyces pombe | Ascomycota | Taphrinomycotina | without | 83 |
| Taphrina betulina | Ascomycota | Taphrinomycotina | forw | 47 |
| Paraphysoderma sedebokerense | Blastocladiomycota | incertae sedis | forw | 66 |
| Allomyces javanicus | Blastocladiomycota | incertae sedis | rev | 161 |
| Geranomyces variabilis | Chytridiomycota | incertae sedis | forw | 68 |
| Zygnema circumcarinatum | Streptophyta | incertae sedis | without | 321 |
| Chlorella vulgaris | Chlorophyta | incertae sedis | rev | 38 |
Table 15: 5S insertions, orientation of the 5S insertions and rDNA copy number per haploid genome of de novo assembled rDNA units.
Figure 5: Phylogenetic dendrogram of de novo assembled rDNAs. Species with names written in red have 5S insertions in the rDNA units, and species with names written in blue lack 5S insertions in the rDNA units.
Congruent with the results of the first part of the analysis, the three Basidiomycota belonging to the Agaricomycotina and the species belonging to the Pucciniomycotina had 5S gene insertions in the rDNA unit, whereas the species from the Ustilaginomycotina did not. Also congrunent results were obtained for the Ascomycota (with the exception of Taphrina betulina), Blastocladiomycota and Chytridiomycota.
The alga Z. circumcarinatum, a member of the higher-plant group Streptophyta, had no 5S insertions. The alga C. vulgaris is a member of the Chlorophyta, which is a sister group to the Streptophyta, and has 5S genes in the rDNA units.
The orientation of 5S insertions is not homogeneous. In three of the analysed Basidiomycota, 5S had the same orientation as the 35S gene, but in Cerinomyces ceraceus, the orientation was reversed. In taxa of the Saccharomycotina, the 5S gene had reversed orientation. Importantly, the orientation of the 5S gene in the two Blastocladiomycota was reversed. The number of repeats ranged from 25 copies in Candida auris to 161 in Allomyces javanicus.
Analysing the rDNA assemblies with the sliding window approach revealed that the rDNA unities within a genome were not necessarely identical. In most of the analysed species, polymorphism were present downstream of the 25S region. The polymorphism was distinct in the yeast species Candida auris, C. parapsilosis and Saccharomyces cerevisiae, but missing in Morchella importuna and Taphrina betulina.
rDNA assembly
Assembling fungal genomes to chromosomal quality is still challenging. Whereas the assembly of greater parts of the genome from NGS data into contigs is now routine, a small portion of the genome is often difficult to assemble, which leads to a higher contig number than number of chromosomes. The rDNA segment is an intricate part of the genome with highly repetitive sequences. The number of rDNA repeated unit’s ranges from 14 to 1442 in fungal species (Lofgren et al.) [16], but only approximately 15% of the analysed assemblies include a complete rDNA unit, and less than 9% include an rDNA array with at least three repeats. In addition to the known obstacles in assembling highly repetitive sequences, rDNA units can have repetitive structures within themselves, regions that are variable between them (James et al. 2009) and intergenic spacers with low complexity. To overcome these challenges, we wrote a program called YNGS allowing the assembly of paired-end fastq data. The assembly starts from a seed and increases the string loop by loop. In the case of an interruption, the fastq fragments, including the paired-end fragments, of the region can be displayed, and the string can be elongated manually. After finishing the string, the sequence is verified by the sliding window approach.
rDNA structure throughout the living organism
Most living organisms have multiple copies of rDNA operons. The copy number in bacterial species commonly ranges between 1 and 15, where more than 80% of the species have more than one copy. The 5S, 16S, and 23S rDNA genes, homologous to 5S, 18S and 25S rDNA in eukaryotes, are organized in gene clusters (Espejo and Plaza) [17]. In archaea, the copy number is generally lower and ranges from 1-4 copies, and the 5S gene is not always part of the operon and can be transcribed separately [18]. As in eukaryotes, both structural variations, in which 5S rDNA is positioned within or outside the rDNA repeat unit, are observed in archaea. However, the rDNA structure of archaea shares more similarity with the rDNA structure of eukaryotes.
rDNA repeats without 5S insertions are common in eukaryotic organisms. Beyond the numerous species in the fungal kingdom, only a few species have been reported to have 5S insertions in their rDNA arrays (Table 16).
| Taxa | Systematic | Reference |
|---|---|---|
| Toxoplasma gondii | Alveolata; Apicomplexa; | Guay15 et al. [42] |
| Dictyostelium discoideum | Amoebozoa; Dictyostelia; | Hofmann et al. 1993; Maizels et al. 1976; |
| Meloidogyne arenaria | Metazoa; Nematoda; | Vahidi et al. 1988; Vahidi et al. 1991; |
| Calanus finmarchicus | Metazoa; Arthropoda; Crustacea; | Drouin et al. [43] |
| Temora longicornis | Metazoa; Arthropoda; Crustacea; | Drouin et al. [43] |
| Semibalanus balanoides | Metazoa; Arthropoda; Crustacea; | Drouin et al. [43] |
| Thysanoessa raschii | Metazoa; Arthropoda; Crustacea; | Drouin et al. [43] |
| Araneus | Metazoa; Arthropoda; Arachnida; | Drouin et al. [43] |
| Chlorella vulgaris | Chlorophyta; Trebouxiophyceae; | this study; |
| Funaria hygrometrica | Streptophyta; Bryophyta; | Capesius 1997; |
| Ginkgo biloba | Streptophyta; Spermatophyta; | Galián 2012; |
| several cryptogams | Streptophyta and Chlorophyta | Wicke et al. 2011 |
| Asteraceae | Streptophyta; Spermatophyta; | Garcia et al. 2010; |
Table 16: Records of 5S insertions in the rDNA arrays of eukaryotic taxa outside the fungal kingdom.
Strikingly, the rearrangement of 5S rDNA occurred several times in the evolution of eukaryotes. By comparing phylogenetic data with rearrangement data of rDNA genes, it is possible to reveal the evolution of rDNA. In land plants, 5S rDNA popped out of the rDNA array at least twice (Wicke et al.) [19]. In early-diverging land plants, rRNA genes were co-localized within the repeated units, whereas most basal seed plants and the distinct group of water ferns had separated 5S genes. Additionally, the opposite rearrangement occurred in the evolution of plants. In the modern family Asteraceae, nearly 25% of the species had a linked arrangement of rRNA genes. These species belong to the tribe Anthemideae, tribe Gnaphalieae and “Heliantheae alliance”, whereas in the other five tribes, only 5S genes outside the rDNA array were detected (Garcia et al.) [20]. The evolution in animals is similar, and 5S rDNA linkages were repeatedly established and lost during the evolution of eukaryotic genomes (Drouin and de Sá) [21].
rDNA structure throughout the fungal kingdom
In fungi, the 18S, 5.8S and 25S rDNA genes are organized in tandem repeats. Tandem repeat arrays can be organized in the genome in one string or separated in a few arrays. Two kinds of arrays can be discriminated. In the first case, each 5S gene is integrated into rDNA units in the intergenic spacer region. In the second case, these insertions are missing, and the 5S genes are scattered within the genome. Then, the 5S genes are typically separated from each other and not arranged in 5S tandem arrays, as is the case in most genomes of higher plants and animals. Most Ascomycota lack 5S insertions in the rDNA repeats, with the exception of Saccharomycetes. Most Basidiomycota have 5S insertions, with the exception of Ustilaginomycetes. For Mucoromycota, Blastocladiomycota and Chytridiomycota, data availability is limited, but it seems that 5S insertions are common in these three phyla. Our analysis and that of Bergeron and Drouin [22] revealed only a very few exceptions to the rules. Some of the exceptions are supposedly artefacts. In particular, when all closely related species are homogeneous and fit the rules, aberrant data should be verified by other methods. In a few groups of Saccharomycetes, aberrant data occur in clusters, which indicate that they are not caused by artefacts.
Ascomycota: The 5S rDNA distribution in Ascomycota fungi can be divided into two groups. The first group included all Ascomycota fungi, without Saccharomycetes, and the second group included Saccharomycetes. For the first group, Ascomycota fungi without Saccharomycetes, Bergeron and Drouin reported two exceptions, Fusarium solani and Pyrenophora graminea, in addition to Cercospora sojina and Taphrina betulina in this study. The few exceptions should be treated with caution, since the results are not stringent and were not verified by a second method. Additionally, we could not confirm the insertion of 5S in the genera Fusarium and Pyrenophora, and a total of 21 Fusarium and 11 Pyrenophora assemblies were analysed in our study.
In non-Saccharomycetes taxa, the 5S rDNA genes were scattered in the genome and are mostly separated from each other. Neighbouring 5S genes were detected very seldomly, whereas the largest array was detected in the genome of B. graminis. In our study, four assemblies of B. graminis were integrated, and in each assembly, 5S gene arrays were detected. B. graminis has an ~120 Mb genome, which is unusually large, e.g., nearly 10 times larger than that of S. cerevisiae. The genome consists of 64% transposable elements, and the dysfunctionality of the RIP pathway has probably contributed to genome-size inflation and to the increase in highly repeated sequences (Spanu et al.) [22,23].
For the Saccharomycetes, 5S insertions in the rDNA units are typical, whereas for the other taxa, 5S insertions are missing. Yarrowia lipolytica within Saccharomycetes is an exception because it lacks a 5S insertion (Bergeron and Drouin) [4,22]. In our study, we detected seven other Saccharomycetes species without 5S insertions: Hyphopichia burtonii, Hyphopichia pseudoburtonii, Komagataella pastoris, Komagataella phaffii, Metschnikowia citriensis, Metschnikowia reukaufii and Sugiyamaella lignohabitans.
Basidiomycota: In the Basidiomycota group, 5S insertions in the rDNA repeats were typical, whereas in the Ustilaginomycetes group, the insertions were missing. Only eight assemblies in the non- Ustilaginomycetes were aberrant, without 5S insertion. Three assemblies in the family Sparassidaceae were aberrant. Bergeron and Drouin reported the loss of 5S insertions in one of two Filobasidiella species. Whereas all analysed Ustilaginomycetes lacked 5S insertions, in other groups of Ustilaginomycotina, 5S insertions were present. Additionally, three Tilletia spp., belonging to the Exobasidiomycetes within the Ustilaginomycotina, were reported to have 5S insertions in rDNA arrays (Bergeron and Drouin) [22].
Blastocladiomycota: In the group of Blastocladiomycota, all analysed taxa had 5S insertions. We analysed the data of two genome assemblies and two additional assembled rDNA repeats from NGS data.
Chytridiomycota: We analysed two taxa within the Chytridiomycota, one genome assembly and one de novo assembly, and in both cases, 5S insertions were present. In contrast, Bergeron and Drouin reported that Batrachochytrium dendrobatidis lacks 5S insertions [22].
Microsporidia: The rDNA organization of Microsporidia deviates from that of other fungi. The 18S and 25S genes are unusually short. The 5.8S rDNA sequences are missing in the ITS between the 18S and 25S genes; instead, in Nosema bombycis, a sequence homologous to the 5.8S gene was detected in the 25S gene. Stringently, in N. bombycis, 25S is located on the reverse DNA strand of the 18S gene (Huang et al.) [24]. Insertions of 5S genes in the rDNA repeat units were detected in three Nosema spp. (Dong et al.) [25,26]. The rDNA genes were repeated in all the genomes analysed so far, and the existence of multiple rDNA arrays was documented for Nosema apis. In Nosema ceranae, significant variations between the rDNA units within a genome were reported, which leads to the conclusion that concerted evolution and homogenization of the rDNA is only rudimentary (Sagastume et al.) [27].
Mucoromycota: For the Mucoromycota group, 5S insertions were identified. In our study, two assemblies were integrated, and in the study of Bergeron and Drouin [22,28], four taxa were studied.
Evolution of rDNA
Whereas the molecular mechanism of 5S gene rearrangement is still unknown, the knowledge about rearrangement of complete rDNA units and copy number variations is much more advanced. The rDNA copy number is strikingly variable in eukaryotes, with 39–19 300 copies in higher animals and 150–26 048 copies in plants. The rDNA copy number in fungi, calculated among 91 fungal taxa, ranged from 14 to 1442. Interestingly, the copy number within a species can be highly variable. For example, among 12 different isolates of Suillus brevipes, the copy number ranged from 72 to 156 (Lofgren et al.). One factor that influences the actual copy number is the environment. The chemical lithium acetate, which is routinely used in the gene transformation of yeast, can change the copy number of rDNA in S. cerevisiae (Kwan et al.) [29]. Salim et al. reported that yeast cells with a reduced number of rDNA repeats grew better than their counterparts with normal copy numbers under conditions of DNA replication stress (Salim et al.) [30]. Another important factor influencing the copy number is genetics. A mutation in the orc4 gene, resulting in a delay of replication initiation and consequently in a prolonged S-phase, severely reduced rDNA copy number. Although reducing rDNA copy number may help ensure complete chromosome replication, orc mutant cells struggle to meet the high demand for ribosomal RNA synthesis (Sanchez et al.) [31].
Despite the fact that the number of rDNA repeats varies, cells strive to keep the number of rDNA repeats in a favorable range. For S. cerevisiae, the average copy number is approximately 150 (Kobayashi et al.). Homologous recombination between the repeated rDNA units led to a reduction in the copy number (Iida and Kobayashi) [32]. The loss of rDNA copies in S. cerevisiae was counteracted by gene amplification, which increased the copy number (Iida and Kobayashi) [33]. The replication fork barrier (RFB) sequence downstream of the 25S gene is important for the reduction and the increase in the copy number (Figure 6). Knocking out the Fob1 protein, which binds to the rDNA replication fork barrier site, disabled a change in rDNA copy number (Kobayashi et al.) [34]. Although knowledge about the mechanism underlying changes in the rDNA copy number is already substantial and sophisticated models exist (Kobayashi, Egidi et al.) [35,36] the mechanism of long-term evolution is still unclear. In the long term, the entire rDNA array could move from one position in the genome to another and even jump to other chromosomes, as detected within members of the family Saccharomycetaceae (Proux-Wéra et al.) [37].
Figure 6: Intergenic spacer region of Saccharomyces cerevisiae. The Intergenic Spacer Region (IGS) is a highly active region for the regulation of rDNA transcription and replication and has a Replication Fork Barrier (RFB), Autonomously Replicating Sequence (rARS), promoters (E-PRO and C-PRO) and nucleosome positions.
5S rDNA is a mobile element that has changed position and orientation several times during evolution. As a result, no continuous lineage from the early evolved eukaryote to modern species exists. On the other hand, we see large groups where the 5S position is constant. The molecular mechanisms of insertion, deletion and inversion of 5S rDNA remain unclear, as do their evolutionary consequences. With the deletion of 5S from the main rDNA array, a reduction in the 5S copy number is normally detected. In species with 5S linked to the rDNA array, the numbers of 5S insertions and rDNA units are equal. In species with separated 5S insertions, the copy number is normally lower, e.g., in S. pombe, with 6 copies. The copies are separated and variable, allowing independent evolution of the 5S sequences. Among animals, including some amphibians, bony fishes and sea urchins, 5S rRNA genes from different loci are expressed in oocytes and somatic cells (Komiya and Bellavia et al.) [38,39].
Molecular mechanism
The exact molecular mechanism underlying the switch from 5S rDNA insertion to deletion and vice versa is unknown. In principle, after insertion or deletion of the 5S gene in one rDNA unit, the new rDNA variation can spread by rDNA rearrangement and homogenization. From the model S. cerevisiae, we learned about the molecular mechanism of rDNA rearrangement, maintaining the rDNA copy number and rDNA homogenization. The rDNA region is highly active, and the intergenic spacer region. Figure 6 performs the main regulatory role in the processes. All three RNA polymerases and the replication machinery are recruited here, and collision of the processes needs to be avoided [40,41].
The replication of the rDNA starts at the autonomously replicating sequence (ARS), and in the beginning, it is bidirectional. In one direction, towards the 5S gene, replication stops at the next RFB, and in the other direction, towards the 35S precursor, it continues until it meets a stalled RFB [42,43].
A stalled fork is frequently converted into a double-strand break (DBS) and repaired by homologous recombination. Due to the repetitive nature of the rDNA sequence, homologous recombination between different rDNA units could occur and could lead to copy number variations and consecutive homogenization of the rDNA array. In S. cerevisiae, as well as in C. auris and C. parapsilosis, the rDNA is only partially homogenized. A small segment in the intergenic spacer region displays high variability within the array. By comparison of the IGS between different S. cerevisiae strains, high variations were found not only among different strains but also within a strain (James et al.). By cloning and subsequent sequencing of the intergenic spacer, variations within a genotype were also detected in the basidiomycete Puccinia striiformis. The phenomenon in which nearly all of the rDNA gene but not the intergenic spacer region is more or less homogeneous within a strain seems to be common in many fungal species.
[Crossref]
Citation: Blechert O, Zhan P (2022) Fungal rDNA Analysis in the NGS Era: Progress, Insights and Challenges. Fungal Genom Biol. 12:202.
Received: 02-Nov-2022, Manuscript No. FGB-22-19117; Editor assigned: 04-Nov-2022, Pre QC No. FGB-22-19117 (PQ); Reviewed: 18-Nov-2022, QC No. FGB-22-19117; Revised: 25-Nov-2022, Manuscript No. FGB-22-19117 (R); Published: 05-Dec-2022 , DOI: 10.35248/2165-8056.22.12.202
Copyright: © 2022 Blechert O, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.