Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2024)Volume 14, Issue 1
Plastic debris in the ocean serves as a stable ground for the formation of a complex ecosystem, termed plastisphere, which includes a variety of organisms from different taxonomic groups. Not much is known about the relationships between the organisms of the plastisphere communities. In this study we describe a novel symbiotic-like interaction between a marine fungus and several species of pennate diatoms on plastic surfaces that were submerged in the water of a Mediterranean Sea marina in Israel. Scanning electron microscope images of the surfaces revealed a network of fungal hyphae with multiple diatom cells attached to them via the side or the tip of their body. Using Deoxyribonucleic Acid (DNA) metabarcoding for the fungal Internal Transcribed Spacer (ITS) barcode locus, we found that the diatom-bearing fungus belongs to the phylum Ascomycota and that it is more abundant on floating plastic surfaces made of Low-Density Polyethylene (LDPE) compared to the denser Polyethylene-Terephthalate (PET) and glass that sinks. We hypothesize that the observed symbiotic-like relationship may have mutual benefits for both parties, including surface-anchoring for the diatoms and nutritional benefits for the fungus that reflects a recent adaptation for life on floating plastic debris.
Marine fungi; Diatoms; Marine plastic debris; Plastisphere; Symbiosis; ITS barcoding; Nanopore minION
Marine plastic debris is now more abundant than ever before with a recent estimate of 82-358 trillion micro particles floating in the world oceans [1]. The plastic surfaces serve as an ideal substrate for the colonization of complex microbial communities forming a man- made ecosystem-the plastisphere, which is distinguished from that of its surrounding water [2]. Most plastic debris in the oceans floats near the surface and benefits from high exposure to sunlight, allowing the establishment of complex communities that include prokaryotic and eukaryotic phototrophs alongside primary consumers, predators and saprotrophs [3].
The plastisphere phototrophs include a variety of pennate diatoms that have been consistently found in high abundances on plastic surfaces in the marine environment and are amongst the first to colonize them [4-8]. The silica skeleton or the ‘frustule’ of the pennate diatoms is made of two bilaterally symmetric or partially symmetric valves. The detailed structure of the diatom’s frustule, as observed by Scanning Electron Microscope (SEM), often allows their taxonomical classification [9]. The diatoms are crucial for the maintenance of the global biochemical cycles, the carbon and silica cycles in particular and are estimated to carry out one-fifth of the earth's photosynthesis [10,11].
Marine fungi are also abundant on plastic surfaces in the ocean, with several strains that have adapted to utilize the plastic as a rich carbon source [12,13]. Using DNA metabarcoding, studies showed the presence of fungi on plastic surfaces either collected from the marine environment or from surfaces that were immersed in it [14-18]. We have previously shown that plastic surfaces that were incubated for one month in the Herzelia marina, Israel, are covered with a biofilm that includes a rich fungal community and often a network of fungal hyphae [19].
In this study, we describe a novel type of symbiotic-like relationship between a fungus and several species of pennate diatoms that were preferentially found on low-density polyethylene plastic surfaces.
Experiment setup and sampling
LDPE bags (20 cm × 18 cm) were incubated for one month in the water (20-30 cm deep) of Herzliya marina (32° 09’ 38.8" N, 34° 47’ 35.0" E), Israel, during Autumn of 2019, 2020, 2021 and spring of 2020. Before sampling, bags were washed thoroughly with filtered (0.2 μm) sterile artificial Foot Sea Water (FSW). For DNA extraction, samples were kept immersed in FSW on ice until processing. For Scanning Electron Microscopy (SEM), samples were transferred to 4% Paraformaldehyde (PFA) and 1% glutaraldehyde fixation solution for 2-5 hours and then washed with 1 × Phosphate Buffered Saline (PBS) and kept in 1:1 ethanol/PBS solution at -20°C until use.
Microscopy (LM and SEM)
For visualization of the live plastisphere flora and fauna and Lacto- Phenol Cotton Blue (LPCB)-stained samples, we used the Nikon eclipse Ci-L microscope with 10x, 20x and 40x objectives. Photos were taken with Nikon DS-Fi3 Complementary Metal Oxide Semiconductor (CMOS) digital camera. For SEM imaging, we followed our previously tested protocol. In brief, fixed samples were dehydrated in graded ethanol series of 50%, 70%, 85% and 95% ethanol (10 min each), followed by 3 × 15 min in 100% ethanol. Dehydrated samples were air-dried for at least 5 hr in a hood and sputter-coated with 10 nm of platinum/gold (Quorum Q150T ES). Samples were visualized and imaged using ultra high-resolution maia 3 FE-SEM (Tescan) in a range of 3-7 kV voltage.
LPCB staining
In order to identify fungal structures (hyphae, sporangia and spores), lactophenol cotton blue dye was used. The dye is based on methyl blue dye with a lactophenol solution (Phenol C6H5OH, lactic acid CH3CH(OH)COOH and glycerol C3H8O3). LPCB dye stains sugar molecules including glucans and chitin that compose the fungal cell wall [20]. Plastic samples were cut to an appropriate size for microscopy and were placed in a petri dish with 2-3 drops of LPCB for 5 mins and then washed with 1 × PBS. Lastly, washed samples were placed on the top of a microscope glass slide and covered with a cover slip.
DNA extraction and amplification of ITS barcode
For molecular identification of the symbiotic fungi, we relied on a short barcoding region within the Internal Transcribed Spacer (ITS) locus that was previously shown to be comprehensive and specific to fungi [21]. For DNA isolation, three plastic pieces (1mm × 1mm) with diatom-baring fungal hyphae were cut under Light Microscope (LM) and placed into a 1% Sodium Dodecyl Sulfate (SDS) buffer. Samples were then lysed in 100 μg/ml Proteinase-K in 1% SDS buffer with ceramic beads for 1 hour at 56°C while mixing thoroughly every 15 minutes. The following steps were performed using QIAamp DNA kit (Qiagen) according to the manufacturer’s instructions.
For Polymerase Chain Reaction (PCR) amplification, ~10 ng of template DNA was used in 50 μl reaction volume. The PCR reactions included the sample DNA, a positive (yeast DNA) and negative (no template) controls. The ITS barcode was amplified using the ITS4 and ITS86 primers in a PCR reaction which was carried out with the following steps: 2 minutes at 94°C, 32 cycles of 30 seconds at 94°C, 30-90 seconds at 45°C to 57°C, 30-90 seconds at 72°C and final extension at 72°C for 5 minutes. PCR product (1 μl) was run in 1% agarose gel (100 volts, 25 min) alongside a PCR Bio IV DNA size marker. The PCR product was cleaned up on magnetic rack with Solid-Phase Reversible Immobilization (SPRI) magnetic beads (Canvax, Spain) following the manufacturer's instructions.
Nanopore minION sequencing
Because we isolated a dirty sample of the symbiotic fungus, ITS metabarcoding approach was used for the taxonomic identification, under the assumption that the target sequence is expected to generate the most abundant PCR product, as was previously demonstrated [22]. The Nanopore MinION sequencing libraries were prepared using the 1D Native barcoding genomic DNA protocol with EXP- NBD 104 and SQK-LSK 109 kits (Oxford Nanopore Technologies). ~200 fmol of purified amplification products were subjected to DNA repair and end-prep using a NEBNext DNA repair mix and NEBNext ultra II end repair/dA-tailing module (New England Biolabs). The library preparation included two ligation steps. In the first step, the multiplexing barcodes were ligated to the amplicons, using T4 Ligase (New England Biolabs) and then equal molarities of the barcoded amplicons were pooled together. In the second step, the MinION adaptor was ligated to the amplicons of the pooled sample. Each step was followed by DNA purification with SPRI magnetic beads. ITS sequencing library was loaded to a new MinION flow cell (R9.4.1) and sequenced. Base-calling for all libraries was done automatically by the MinKNOW program using the “high accuracy” base-caller option. Raw reads were obtained in FAST5 and formats from which pass quality reads were subjected to further analysis.
Data analysis
The raw nanopore reads were demultiplexed and trimmed with qcat. Primers were removed using Cutadapt with 10% error tolerance. The reads were filtered based on read quality and read length (300- 340bp), based on the plotted quality/length distribution. The filtered reads were used to create a single consensus sequence using the ONTrack pipeline. The consensus sequence was deposited in the genebank (accession no. OQ843024) and was compared with NCBI and Silva databases for taxonomic identification. In order to assess the relative abundance of the symbiotic fungi, the consensus sequence was compared to an ITS database that was created from samples that were incubated in the same marina location for similar time periods. The comparison was conducted with local blast+suite (Version 2.13.0) provided by the National Center for Biotechnology Information (NCBI), wherein a threshold of 80% query coverage and 94% sequence identity were applied. Prism software (v 8.0.2) was used to evaluate significant differences with Holm-Sidak's multiple comparisons test and to create the graphs.
Interactions of diatoms with fungal hyphae: LDPE plastic surfaces that were incubated for one month during spring and autumn of 2019 and 2020 in the Herzliya marina (Israel) contained a rich biofilm that could be noticed by the naked eye. Light and Electron Microscope (SEM) images revealed a network of fungal hyphae with a few fruiting bodies (Figures 1A-1H and 1J-1O). The plastic surfaces that were collected in the 2019 and 2020 spring seasons, contained fungal hyphae with diatoms attached to them. The fungi-attached diatoms were of different species belonging to several pennate diatom genera including Striatellaceae (Figure 1B), Licmophora (Figures 1B and 1N), Nitzschia (Figures 1D, 1H and 1L), Amphora (Figures 1K and 1L) and Achnathes (Figure 1M). The diatom-baring hyphae were positive to lactophenol cotton blue staining (Figure 1C), confirming the presence of typical fungal cell walls. Both fungus-bound and free diatom cells were often found during/after a single binary division (Figures 1C and 1H) or as cell bundles after multiple divisions (Figures 1D and 1N). Dividing diatom cells were found to be attached from one of the frustule’s poles, while non-dividing individuals were attached from their perpendicular plane in all observed cases. The fungus - diatom contact points were enriched by unknown extracellular material, which presumably strengthened the anchoring of the diatom cells to the hyphae filaments.
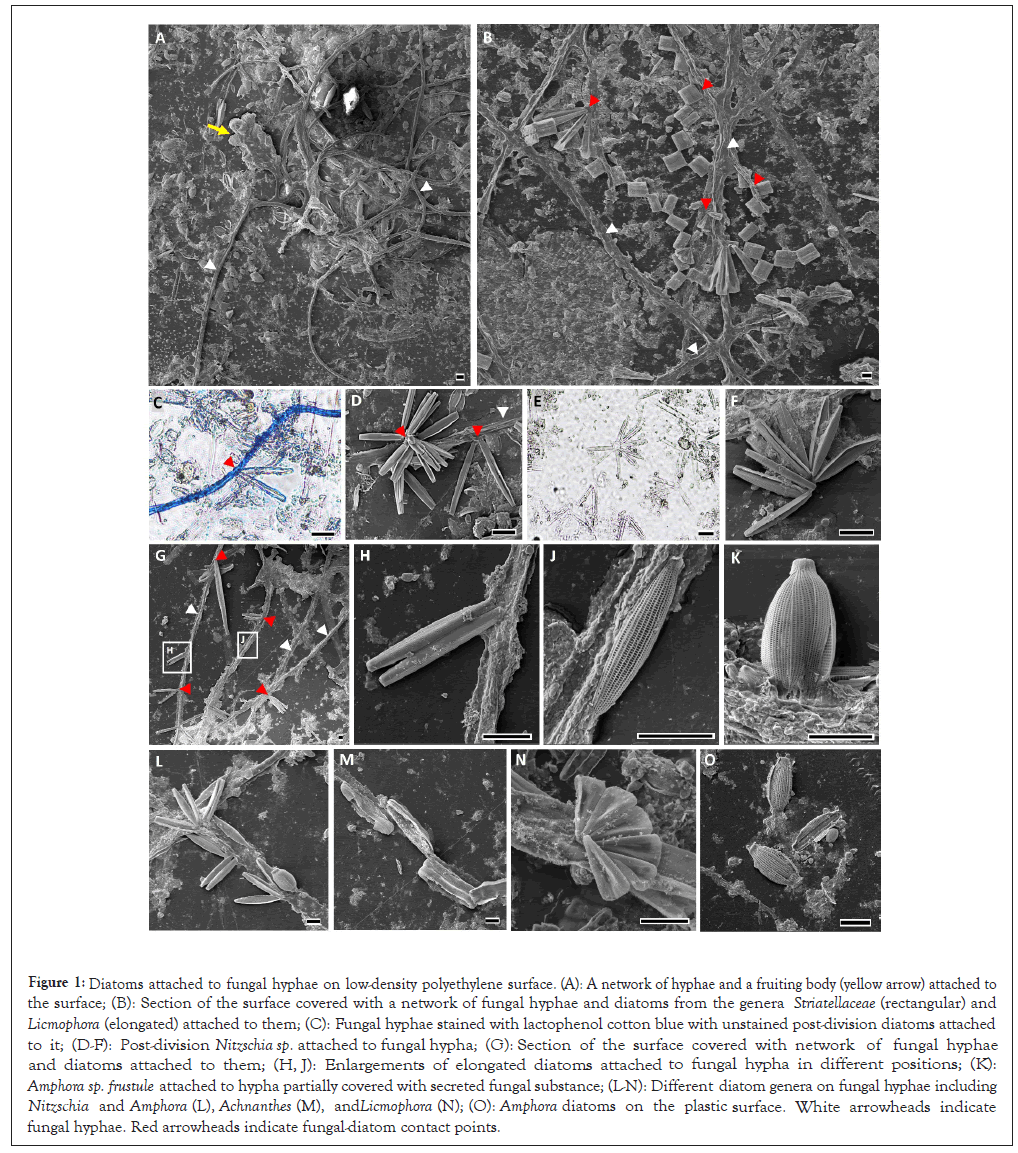
Figure 1: Diatoms attached to fungal hyphae on low-density polyethylene surface. (A): A network of hyphae and a fruiting body (yellow arrow) attached to the surface; (B): Section of the surface covered with a network of fungal hyphae and diatoms from the genera Striatellaceae (rectangular) and Licmophora (elongated) attached to them; (C): Fungal hyphae stained with lactophenol cotton blue with unstained post-division diatoms attached to it; (D-F): Post-division Nitzschia sp. attached to fungal hypha; (G): Section of the surface covered with network of fungal hyphae and diatoms attached to them; (H, J): Enlargements of elongated diatoms attached to fungal hypha in different positions; (K): Amphora sp. frustule attached to hypha partially covered with secreted fungal substance; (L-N): Different diatom genera on fungal hyphae including Nitzschia and Amphora (L), Achnanthes (M), and Licmophora (N); (O): Amphora diatoms on the plastic surface. White arrowheads indicate fungal hyphae. Red arrowheads indicate fungal-diatom contact points.
The prevalence and nature of the fungal-algal relationships: As fungal - diatom firm attachment was repeatedly observed by LM and SEM on LDPE surfaces in what seems like a symbiotic relationship. The average number of diatoms that were found in association with fungal hyphae on 1 × 1cm surfaces was 30 (n=5, STDV=8.02). To define whether the relationships are obligatory or facultative, we searched for diatoms of the same taxa that were not attached to fungi elsewhere on the plastic surface. Indeed, similarly-looking unassociated diatom cells (both single and post-division) were found, indicating that the relationships are likely not obligatory. The most common fungus-bound diatom of genus Nitzschia (Figures 1D and 1L) which accounted of ~48% of the hyphae-bound diatoms (Figures 1A-1H and Figures 1J-1O).
Diatom-baring hyphae belong to an unknown Ascomycota fungus with a preference for PE surfaces: While diatoms may often be classified using high-resolution microscopy images up to the genus and even to the species level, this is not the case for marine fungi. We therefore implemented the DNA barcoding approach to classified the diatom-bearing fungus based on ITS4 and ITS86 primer set for the ITS2 region that amplify an ITS sequence that is regarded as a gold standard barcode for fungal taxonomical identification and outperform the other ITS barcodes. Although DNA was extracted from a small piece of fungus-containing surface, it was reasonable to assume that it was contaminated with spores carrying contaminating fungal DNA. This notion was supported by the large PCR amplicon size range which was observed on the gel image (Figure 2A) and in the analysis of the amplicon read sizes (Figure 1B). We therefore used nanopore MinION high throughput sequencing to identify the most prevalent and repetitive ITS barcode sequence. To reduce background noise, analysis was performed on a restricted sequence length range based on the amplicon size distribution (Figure 2B). The results showed a single consensus of 284 bp (Acc. OQ843024) which was mapped to an unknown taxon from phylum Ascomycota (referred to as fungus “F1”). We further used the consensus sequence to analyse the F1 prevalence in our previously published ITS metabarcoding database. This database was established using nanopore sequencing of the microbiome ITS amplicons, obtained from plastic and glass surfaces that were submerged in Herzliya marina for one-month period in similar to the surfaces on which the fungal-diatom interactions were observed. The results suggest that the F1 fungus was more abundant on surfaces vs. water with a significant preference for floating LDPE surfaces over polyethylene terephthalate (PET) and glass surfaces (Figure 2C).
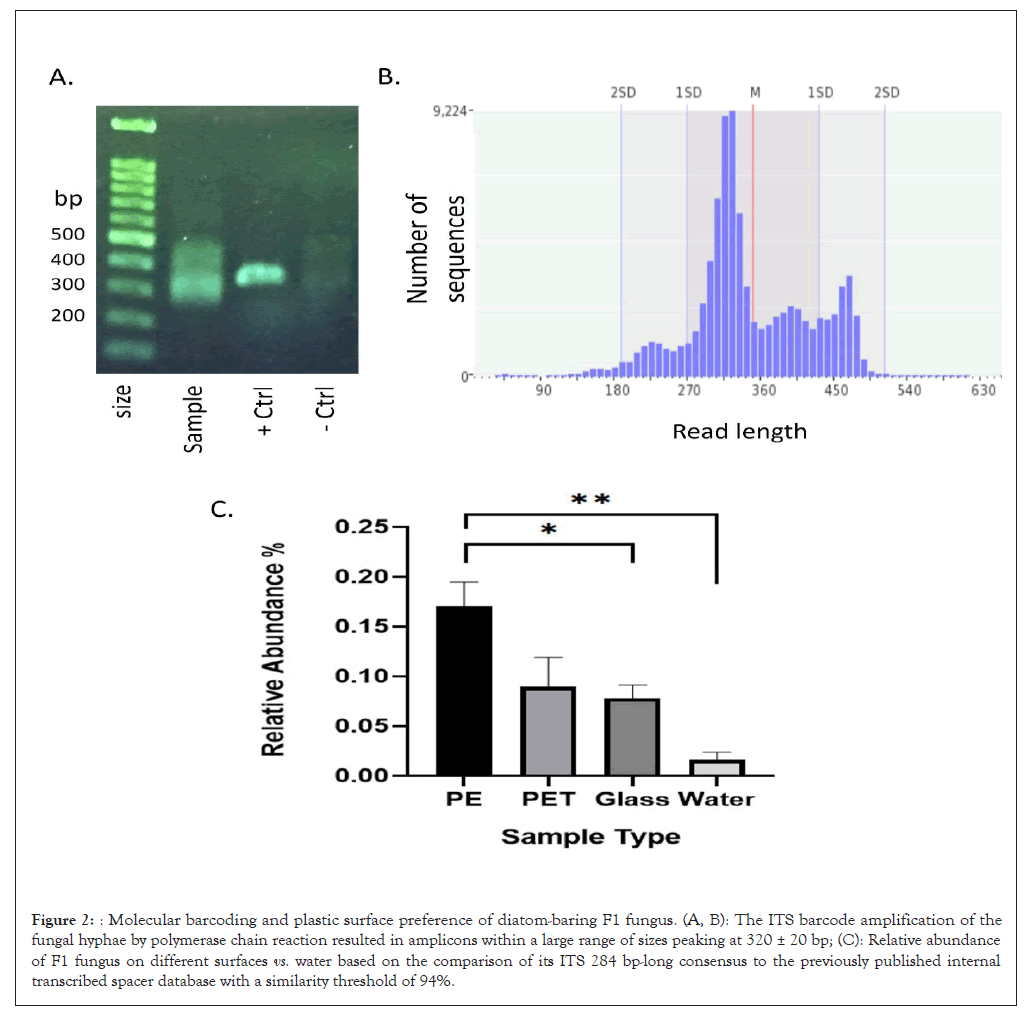
Figure 2: Molecular barcoding and plastic surface preference of diatom-baring F1 fungus. (A, B): The ITS barcode amplification of the fungal hyphae by polymerase chain reaction resulted in amplicons within a large range of sizes peaking at 320 ± 20 bp; (C): Relative abundance of F1 fungus on different surfaces vs. water based on the comparison of its ITS 284 bp-long consensus to the previously published internal transcribed spacer database with a similarity threshold of 94%.
In this study, we characterize a novel relationship between fungi and several species of pennate diatoms on a plastic surface that was submerged in the water of a Mediterranean Sea marina for a period of one month. In a previous study, we identified a network of fungal hyphae that partially covered plastic surfaces that were submerged in the marina. As we did not identify a similar network on plastic debris that was collected in the open sea, we assume that the relatively undisturbed, yet polluted (with organic waste) marina water contribute to the development of such networks. The fungal-diatom interactions were observed in the autumns of three consecutive years (2019-2021) but was not found in spring. This seasonality is not surprising given the overall taxonomic seasonality observed within the plastic biome. Furthermore, according to our ITS barcode analysis and the comparisons to our previously established database, we concluded that the symbiotic fungus is more prevalent on PE surfaces vs. PET or glass surfaces. PE is generally less dense than seawater, therefore tends to float near the water surface, while PET and glass sinks. Thus, the observed preference may reflect an adaption for colonizing surfaces that allow photosynthesis by the photobiont (the diatoms). While we did not investigate the mechanism of the interaction, we suggest that both the fungus and the diatoms gain benefits from it. We suggest that the diatoms use the fungal hyphae as an anchoring network to the floating surface. This notion is supported by the unknown secreted fungal substance, observed by SEM, which seems to glue the diatoms to the hyphae filaments. On the other hand, similar to lichens, the fungus may utilize sugars secreted by the diatoms or even diatom corps (that were observed attached to hyphae).
As marine plastic surfaces are relatively new in our world, it is possible that the observed plastic-associated symbiosis-like relationship were adapted from a preexisting symbiosis that occurs in either shallow benthic marine environments, natural floating organic surfaces such as wood and leaves or even on living organisms such as multicellular algae. This anchoring may allow frequent diatom cell division without losing grip of the surface as was often observed. Our finding presents a new type of relationships that is hosted in the plastisphere ecosystem and that is specifically fungal-algal growth on floating plastics.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Itzahri S, Davidov K, Oren M (2024) Fungal-Diatom Relationships on Floating Plastic Surfaces in the Mediterranean Sea. Fungal Genom Biol. 14:237.
Received: 26-Dec-2023, Manuscript No. FGB-23-28659; Editor assigned: 28-Dec-2023, Pre QC No. FGB-23-28659 (PQ); Reviewed: 11-Jan-2024, QC No. FGB-23-28659; Revised: 18-Jan-2024, Manuscript No. FGB-23-28659 (R); Published: 25-Jan-2024 , DOI: 10.35248/2165-8056.24.14.237
Copyright: © 2024 Itzahri S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.