Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2020)
Background: Alzheimer’s disease (AD) is the leading neurodegenerative disorder affecting memory, learning and behavior. Altered expression of proteins involved in neuronal structure and function is a recent observation of AD pathogenesis. Modulation of altered protein expression seems promising in AD therapeutics. In the present experiment, AD ameliorating effect of medicinal mushroom Ganoderma lucidum (GL) had been evaluated through its effect on neuronal cytoskeletal structure and function related protein expression pattern in AD model rats.
Methods: Wistar male rats (120 ± 5 gm) were divided into three groups: control rat (CR), Alzheimer’s Disease Model Rat (ADMR) and G. lucidum Hot Water Extract (GHWE) fed ADMR, each group containing 15 rats. AD model rats were prepared by infusing Aβ1-42 (ab120959, abcam, USA) into the cerebral ventricles. Protein extraction from the brain samples was performed following homogenization of the hippocampus (50 mg) with lysis buffer (1 ml) using a homogenizer (Polytron PT 1200, Kinematica). Protein separation through SDS-PAGE and protein quantification through LC-chip MS/MS Q-TOF had been performed for label-free relative quantification. For statistical analyses, the data were exported to the Mass Profiler Professional (MPP) software and ANOVA (P<0.05) had been performed to overcome the complications of false discovery associated with multiple test analyses. Functional interaction networks of the proteins were identified using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (version 10.0; http://string-db.org/). For further identifying over-representing pathways and biological functions, the Ingenuity Pathway Analysis (IPA), build version: 389077M, content version: 27821452, (Release date: 2016-06-14) was used (https://www.ingenuity.com/wp-content/themes/ingenuity-qiagen). Datasets of the proteins significantly expressed (P<0.05) and having log FC (fold change) of 1.5 and higher were uploaded (ADMR versus CR, ADMR versus GHWE fed ADMR and CR versus GHWE fed ADMR).
Results: Among 2,212 proteins identified in the present study, 819 had been found to be differentially expressed. Of the differentially expressed ones, 9 proteins had been linked with neuronal cytoskeletal structure and function regulation such as tubulin, β-actin, dihydropyrimidinase-related protein 2 (DRP-2), keratin, Glial Fibrillary Acidic Protein (GFAP), Rho A proteins, septin, cofilin, gelsolin and dynamin. AD rats manifested altered expression of proteins associated with neuronal structure and function. G. lucidum Hot Water Treatment ameliorated the altered expression of those proteins.
Conclusion: Altered expression of hippocampal proteins is a hallmark of AD. Neuroproteomics regulatory approach towards AD amelioration seems promising. Inclusion of G. lucidum for proteomics based AD therapeutics in regulation of the proteins involved in neuronal structure and function seem apt. Thus, G. lucidum could be considered as an AD therapeutic agent species.
Alzheimer’s disease; Neuroproteomics; Ingenuity pathway analysis; Protein-protein interaction; String
AD: Alzheimer’s Disease; ADMR: Alzheimer’s Disease Model Rat; CR: Control Rat; GL: Ganoderma Lucidum; HWE: Hot Water Extract; FC: Fold Change; GHWE: Ganoderma lucidum Hot Water Extract; KDa: IPA: Ingenuity Pathway Analysis; PPI: Protein-Protein Interaction; STRING: Search Tool for the Retrieval of Interacting Genes
According to United Nations Education Scientific and Cultural Organization (UNESCO), mushroom had included in the Ben Cao Gang Mu, the first pharmacopoeaia in China, compiled during Ming dynasty (1590 A.D.) as an aesthetic, spiritual and therapeutic agent [1,2]. Medicinal mushroom, Ling zhi (Ganoderma lucidum) has 2000 years-old reputation of usage in the Chinese medicine [3]. The State Pharmacopoeia of the People’s Republic of China (2000) has incorporated G. lucidum as the potent replenisher of Qi (life force), salient mind soothening and cough relieving as well as anti-asthmatic agent [2]. Modern medicinal sector also acknowledge its inclusion as a therapeutic agent against numerous physiological disorders including, but not limited to, antioxidant, anti-inflammatory, antitumor, anticancer, antimicrobial, immunomodulatory and hepatoprotective agent [3-7]. Content of more than 400 ganocomponents had accredited Ganoderma as the “fungal biofactory”, “panacea” and the “elixir of life” [3-7]. Ganocomponents conferring medicinal values range from polysaccharides to triterpenes, sterols, proteins, peptides, fatty acids and vitamins [3-7]. Recently, utilization of G. lucidum in ameliorating neurodegenerative diseases like Alzheimer’s disease (AD), have received epoch-making attention [8]. We have also observed promising effect of G. lucidum as an AD therapeutic agent [9-15].
AD is a neurodegenerative disorder affecting mostly the elderly people is posing threat to the ever increasing aged humanity of the world. Currently, more than 46 million people over the world have been suffering from AD and this number had been projected to double by 2050 [16,17]. AD patients suffer from progressive loss of memory and learning abilities, behavioral abnormalities, disorientation about time and space, inability to smell and taste, difficulty in performing errands [17]. At severe stage, they become solely dependent on their family members and care-givers that lays extra economic burden on national and global health-care budget. Though the number of AD patients is skyrocketing, the world still awaits successful medication against this age-onset malady. AD occurs due to loss of neurons and synapses associated with memory, learning and behavioral performances [17]. Since its identification in 1901, different hypotheses relating AD pathogenesis had been put forward. Among them, genetic predisposition, formation of amyloid beta (Aβ) plaques, Neurofibrillary Tangles (NFT) and mutation had received attention [14]. Recently, proteomics approaches towards understanding the mechanism and modulation of AD pathogenesis has received high attention [10,18-21]. Differential expression of proteins between AD and normal subjects would aid in formulating therapeutic strategies against AD. Though different approaches of AD therapeutics had been linked with G. lucidum, there is scarcity of reports describing AD neuroproteomics modulating effect of G. lucidum. Thus, the present study had been designed to evaluate the AD neuroproteomics modulatory effect of G. lucidum, especially the expression pattern of proteins involved in maintaining neuronal cytoskeletal structure and function.
Animals
Wistar male rats (120 ± 5 gm) were divided into three groups: CR, ADMR and GHWE fed ADMR, each group containing 15 rats. AD model rats were prepared by infusing Aβ1-42 (ab120959, abcam, USA) into the cerebral ventricles following an established method [22]. All the experimental protocols had been approved by the ethical permission committee, University of Malaya Institutional Animal Care and Use Committee (UMIACUC) [Ethics reference no. ISB/25/04/2013/NA (R)].
Brain sample preparation and protein quantification
Following our previously established method [10], rat brain samples had been prepared and protein quantified in the hippocampi of CR, ADMR and GHWE fed ADMR rats. Briefly, protein extraction from the hippocampi had been performed by homogenization (brain sample 50 mg; lysis buffer 1 ml; homogenizer Polytron PT 1200, Kinematica); protein separation had been performed through SDS-PAGE [mini- PROTEAN tetra cell (165-8000, BIO-RAD, USA)] and protein quantification through LC-chip MS/MS Q-TOF [10]. To identify the proteins, the acquired MS/MS data were compared against the UniProtKB/Swiss Prot rat (Rattus norvegicus) database using the Spectrum Mill and X! Tandem. The differentially expressed proteins in the different groups were identified using their canonical sequence and proteins having fold change of at least 1.5 times were considered as the deregulated proteins. For validation of the identified proteins, the data were exported to the Scaffold database (version 4.5.1, Portland, USA). Proteins were grouped together if they would share at least two peptides and maintained their threshold level at 95.0% and <1% False Discovery Rate (FDR) by the Peptide Prophet algorithm with Scaffold delta-mass correction for the matched peptide-spectra.
Statistical analysis
Data had been exported to the Mass Profiler Professional (MPP) software that analyzed depending on the MPP entities, the intensity of the total spectra of the proteins. Analysis of Variance (ANOVA) had been performed to overcome the complications of false discovery associated with multiple test analyses.
Bioinformatics and analysis of protein-protein interaction
Most of the proteins do not work singly rather they participate in complex network or scaffold and interact with others. Thus, analysis of the relevant protein-protein networks provides important information in deciphering any bio-molecular system. Functional interaction networks of the proteins were identified using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database. STRING displays protein-protein interactions in a large network of connectivity and protein hubs. Active prediction methods that we used were experiments, neighborhood, databases, gene fusions, coexpression, cooccurrence and text mining, using high confidence (0.7).
For further identifying over-representing pathways and biological functions, the ingenuity pathway analysis (IPA), build version: 389077M, content version: 27821452, (Release date: 2016-06-14) was used. Datasets of the proteins significantly expressed (P<0.05) and having log fold change of 1.5 and higher were uploaded (ADMR versus CR, ADMR versus GHWE fed ADMR and CR versus GHWE fed ADMR).
Differential expression of proteins involved in neuronal cytoskeleton maintenance
Differential expression of the proteins involved in maintaining neuronal cytoskeletal structure and function had been observed in the present study. Derangement of neuronal cytoskeleton through microtubule disassembly is an important feature of neurodegeneration [23]. STRING analysis revealed strong networks among the microtubule assembling cytoskeletal proteins such as tubulin, β-actin, dihydropyrimidinase-related protein 2 (DRP-2), keratin, Glial Fibrillary Acidic Protein (GFAP), Rho A proteins, septin, cofilin, gelsolin and dynamin.
Tubulin
AD neurons suffer from disrupted microtubule structure and functioning [24]. Tubulin is the main component of microtubule and consists of dimers imparted by the alpha and beta chains. Differential expression of tubulin α -1c, -4a, β -2a, -2B, -3 and -5 chains were observed in the present study. Molecular function based sub-network analysis showed different tubulin chains to be clustered together and deranged in AD. Both animal and human studies have linked decreased level of α and β tubulin with human AD [25,26]. In AD brain, β tubulin becomes abnormally hyperphosphorylated and modified tubulin fails to assemble microtubules. Consequently, microtubule disassemblage leads towards cytoskeletal vulnerability [27]. Recently, microtubular disassembly has been implicated in causing “mitochondrial traffic jam” in the AD neurons as mitochondrial shifting across the “rail-road of microtubule” becomes impeded in the AD brain [28].
β-actin
Normally, β-actin is involved in maintenance of cytoskeleton, internal cell motility, neuronal network integrity and aids in memory and learning performances. Its altered expression and oxidized form had been linked with AD pathogenesis [29]. Impaired expression of actin is in agreement with the synaptic dysregulation associated with AD and age-related altered cytoskeletal structure, axonal dystrophy, reduced dendritic spines and impaired transport across membranes [30]. Enhanced accumulation of actin enhances tau-governed neurotoxicity [31,32].
Dihydropyrimidinase-related protein 2 (Drp-2)
Dihydropyrimidinase Related Protein 2 (DRP2) is involved in regulation of axonal outgrowth and becomes hyperphosphorylated in NFT and its increased level is observed in AD model animals [33]. Compared to the normal neurons, AD neurons possess shortened dendrites which are a characteristic of their lowered communication with neighboring neurons [34]. DRP2 is expressed highly in the developing brains and altered in AD [35,36]. It regulates the activity of collapsin that elongates the dendrites, increases their communicability and repairs the damaged neurons [35,36]. Oxidative modification of DRP2 might cause reduced length of the dendrites and communication leading to lowered cognitive performance [37].
Glial fibrillary acidic protein (Gfap)
Glial Fibrillary Acidic Protein (GFAP) provides structural support to the astrocytes and its elevated level in AD model animals and in human subjects [33,38]. Its differential and reduced expression has been reported in AD animal studies [39,40].
RhoA proteins
Ras homolog gene family, member A (RhoA) proteins are involved in cytoskeleton regulatory processes such as dendrite development, axonal extension and protrusion [41]. They also stabilize the Aβ-disrupted microtubules [41]. Aβ increases RhoAGTPases and decreases neuronal spine production and neural connection both in the cell lines and also in the brains of the transgenic AD models [42,43]. Inhibition of the RhoA/ROCK signaling pathway seems ameliorating towards axonal growth and cognitive decline [41].
Septin
Septins are microtubule associated, filament-forming and GTPbinding proteins that participate in dendritic spine formation and in neurotransmitter release [44,45]. Like that of Musunury et al. [46], increased expression of septin-2 and -3 were noted in the AD rats’ hippocampi. Its increased expression in the AD rats might be involved in disrupting microtubular filament formation and associated cytoskeletal derangement in the AD rats [47-49].
Cofilin
Brain cofilin activity reduces with age and in the AD subjects, it goes down aberrantly [50]. Transgenic mice studies also showed its lowered expression [50]. However, unlike other cytoskeletal proteins, its levels have not been found to be up-regulated in AD ameliorating cases [51]. As cofilin is a regulator of actin, increased cofilin expression points towards increased actin turnover and increased depolymerization of actin filament [51].
Dynamin
Dynamin is a neuronal GTPase capable of free entry into and release from the synaptic vesicles [31]. Current finding of its decreased expression in the AD rats is consistent with those of others [52-55]. Aβ-induced depleted dynamin 1 level has been found to impair memory in the AD model rats [53]. Although some other findings found its variable level, altered dynamin induced affected neurotransmitter release has commonly been correlated with AD pathogenesis [32,55].
Gelsolin
Gelsolin is a member of the actin-binding proteins having antioxidative, Aβ binding and fibrillation inhibitory potentiality [56-58]. Its overproduction and/or administration showed Aβ lowering effect and thus, gelsolin has been regarded as an AD therapeutic agent [59]. More importantly, administration or overexpression of gelsolin results in significant reduction of amyloid load and decrease of Aβ level in AD transgenic mice [56,59]. Down-regulated expression of gelsolin was noted in the AD rats’ hippocampus that is in par with those of Manavalan et al. [60]. However, some studies have reported confounding results regarding the expression of gelsolin in AD brains [61].
PPI network of the upregulated protein clusters
The upregulated proteins constituted the interaction network entailing microtubule assembly and cytoskeletal structure and function (Figure 1). The proteins involved in this network are sirtuin 2 and tubulin with the corresponding genes Sirt2, Tuba1b, Tuba1c, Tubb2a, Tubb3, Tubb2b, Tuba4a, Tubb4a, Tubb4b, Tubb5 and Tubb6 (Figure 1).
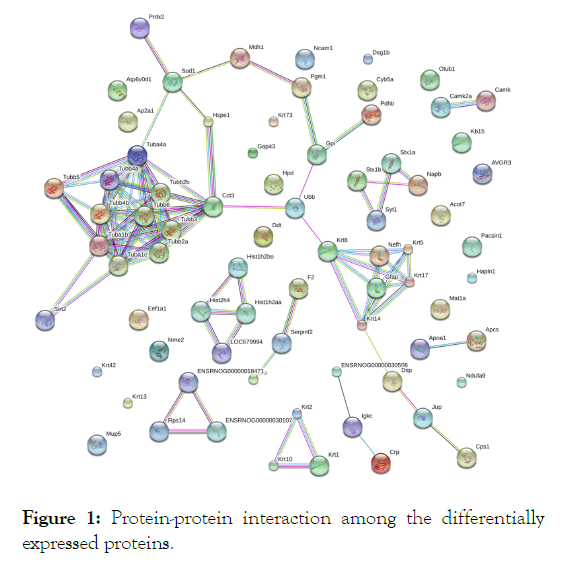
Figure 1: Protein-protein interaction among the differentially expressed proteins.
An important feature of the present study is that some of the proteins differentially expressed in the AD model rats are highly interacted with each other and formed pathway-based functional networks. This inter-relationship sets a novel ground for AD therapeutics. Thus, findings of the current research suggest a novel target for AD pathogenesis management and incorporation of G. lucidum as an AD ameliorating agent seems pertinent. Differential expression of the proteins in the mushroom-fed AD rats might either be an adaptive response or protective strategy against Aβ-mediated stresses. Admittedly, the regulated proteins, identified in the current experiment but not previously been studied, warrants extensive exploration for much conclusive remarks. In this endeavor, western blot analysis of the mostly regulated proteins for validation of the current findings is the immediate future aspect of the present study.
Present study indicates the Alzheimer’s disease modulatory effect of medicinal mushroom G. lucidum through differential expression of neuronal cytoskeleton regulatory proteins, restoration of disrupted protein-protein interaction network and maintenance of integrated pathways. Thus, utilization of G. lucidum in AD therapeutics seems promising. However, further studies should be carried out to determine therapeutic dosage, toxicity and safety.
Ethics approval and consent to participate
All the experimental protocols had been approved by the ethical permission committee, University of Malaya Institutional Animal Care and Use Committee (UMIACUC) [Ethics reference no. ISB/25/04/2013/NA (R)].
Consent for publication.
All authors consented to publish.
Availability of data and material
Not applicable.
Authors gratefully acknowledge the financial support from Jahangirnagar University, Bangladesh; University Grants Commission (UGC) of Bangladesh; Bangladesh Ministry of Education; Ministry of National Science, Communication and Information Technology of Bangladesh and University of Malaya, Malaysia.
Mohammad Azizur Rahman designed and conducted the experiments, interpreted outcomes, composed manuscripts. Other authors supervised the experiments.
Authors gratefully acknowledge the personnels and institutions that provided laboratory and equipmental support in conducting this research.
Citation: Rahman MA, Hossain S, Abdullah N, Aminudin N (2020) Ganoderma lucidum Modulates Neuronal Distorted Cytoskeletal Proteomics and Protein-Protein Interaction in Alzheimer's Disease Model Animals. Fungal Genom Biol. S1: 002.
Received: 17-Nov-2020 Accepted: 03-Dec-2020 Published: 10-Dec-2020 , DOI: 10.35248/2165-8056.20.S1.002
Copyright: © 2020 Rahman MA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.