Entomology, Ornithology & Herpetology: Current Research
Open Access
ISSN: 2161-0983
ISSN: 2161-0983
Research Article - (2019)Volume 8, Issue 2
Maintenance of intraspecific polymorphisms is important for biodiversity. The aphid Macrosiphoniella yomogicola has two color morphs (green and red) in Hokkaido. Their primary attending ants (Lasius japonicus) can manipulate the frequency of the green morph to match that of the red morph to maintain the polymorphism in an aphid colony. Ants prefer the green morph owing to its high-quality honeydew; however, the ants intentionally maintain nutritionally inferior red morphs in every colony because the ants only manipulate the reproductive rate of the green morph, i.e., the ants discriminate between the two morphs. Thus, the benefit of the red morphs to the ants should differ from nutrient exploitation. This hypothesis requires a genetic trade-off between the two morphs. Here based on three microsatellites, we show that the genetic distance between both morphs collected from the same sampling site is small suggesting that there is no genetic differentiation between their whole genomes. However, there were significant differences between the two morphs at a locus at the three sites examined. Our results suggest that intermorph copulations occur; however, the loci controlling morph-specific traits should be linked with the differentiated locus.
Maintenance of genetic polymorphisms is important for understanding the emergence and maintenance of biodiversity in nature [1,2]. Four main hypotheses may explain this phenomenon: 1) different adaptations of each allele in a gene pool to different patches in a homogeneous environment [3,4]; 2) frequencydependent selection among multiple alleles [5-8]; 3) heterosis; and 4) in a symbiotic system, manipulations by one species that neutralize reproductive competition among genetic morphs of the other species [9,10]. In particular, there have been few studies investigating hypothesis 4, although a symbiotic system requires and simultaneously maintains biodiversity. Thus, the roles of symbiotic systems in understanding biodiversity should be elucidated further.
In the myrmecohillous aphid Macrosiphoniella yomogicola, there is a color polymorphism [11]. In Hokkaido, the morphs can be classified into two types, green and red [12]. When the aphids are not attended by ants, there is a higher increasing rate of the red morph compared to that of the green morph [9,10]. However, when ants attend the aphids, the frequencies of the red and green morphs are similar, indicating manipulation by the ants to neutralize reproductive competition between the two aphid morphs [9,10].
Additionally, the attending ants prefer the green morph under both experimental and field conditions [10], likely owing to the highquality honeydew excreted by the green morph. If this observation is correct, although the ants can discriminate between the morphs since they manipulate the green morph only, why is the red morph maintained in aphid colonies by the ants? In fact, the attending ant Lasius japonicus has been reported to remove individuals of another symbiotic aphid for inferior honeydew excretion [13], (here L. japonicus was classified as L. niger). A reasonable explanation for the maintenance of the red morph is it has a different characteristic that is advantageous for the ants, and thus, L. japonicus benefits from maintaining the red morph. However, if this is the case, there should be a genetic trade-off between the beneficial traits of both color morphs. If there is not such a trade-off, a “super morph” with the beneficial traits of both the red and green morphs should be dominate in the populations.
In this study, we analyzed the genetic structure within and among three populations of M. yomogicola using three microsatellites, focusing, in particular, on the genetic differentiation between the color morphs. Based on the results, we discuss the above possibility, namely that the inevitable genetic trade-offs between genotypes maintains biodiversity in a symbiotic system.
Materials
The aphid Macrosiphoniella yomogicola inhabits mugwort, Artemisia montana. A. montana forms a genet with multiple clonal individuals connected by the root (each individual plant is called a shoot). M. yomogicola has two color morphs (green and red) in Hokkaido [12]. In mid-May, stem mothers appear from overwintered eggs that are produced by sexual reproduction in the previous late fall. They climb to the top of the shoot that they inhabit and begin producing female offspring by asexual reproduction. Soon after the habit of stem mothers, attending ants arrive and care for the aphids to exploit their honeydew. In late June, winged forms of both morphs appear, and they disperse to other shoots. From late June to early August, most of the mugwort shoots are inhabited by the mixed morph aphid colonies. After inflorescences of the host shoot have emerged, aphid colonies rapidly decrease [10]. When an aphid colony survives until mid-October when sexuparae of M. yomogicola emerge, inhabited shoots rarely produce inflorescences. Then, sexual females lay overwintering eggs after copulation. Mating between green morphs (G) and red morphs (R) has been confirmed in the field [14].
Aphids could not survive without attending ants [12], and ants prefer G to R [9]. R increase more rapidly than G in the absence of ants, but ants neutralize this disparity between R and G by equalizing the rate of increase of both morphs by improving the increase rates of G to be similar to those of R [9]. Thus, although attending ants can discriminate between G and R, they intentionally maintain R in the colony. A potential hypothesis explaining why both morphs are retained is R provides another benefit from G to the ants, and thus maintaining both morphs is beneficial for the ants. If this hypothesis is correct, there should be a genetic trade-off between the abilities of the morphs to benefit the ants. If the hypothesis is not supported, a super morph with both beneficial abilities should be selected for and should be dominant in the populations (i.e., the color polymorphism should disappear).
Sample collection
On July 26, 2018, we collected five shoots of the host plant, Artemisia montana, which had been parasitized by M. yomogicola, from three sites (JA: the side of the parking area of the Faculty of Veterinary Medicine within the property of Hokkaido University, JB: the other side of the parking area of the Faculty of Veterinary Medicine, and En: near the School of Engineering). The linear distances between the collection sites were measured using ImageJ (ver. 1.51h) and were as follows: JA to JB=41.4 m, JA to En=705.9 m, and JB to En=674.5 m. From each shoot, eight red and eight green individuals were sampled. When there were fewer than eight individuals for a morph, we used all individuals of that morph.
Preparation of microsatellites
Microsatellites were designed following the methods described [15] resulting in primer-pairs for 18 loci. As an initial screen for polymorphic loci, we made primers without fluorescent labels and amplified a mixture of total DNA from 20 M. yomogicola individuals from various shoots using 28 cycles of Polymerase Chain Reaction (PCR) consisting of 30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C followed by 7 min at 94°C. Then, the amplified products of each locus were separately electrophoresed in a 1.0-mm thick 1.5% acrylamide gel in 1.0% Tris-borate buffer at 200 V for 2 hrs. Gels were stained for 20 min in 250 ml of 0.2% ethidium bromide and were examined for multiple bands of approximately the target length. When multiple bands were observed that were approximately the target length, we made a fluorescent label for one primer of the primer-pair. Additionally, we amplified five loci in the same genus to identify polymorphisms [16]. Allelic polymorphism at each locus was examined by amplifying DNA from eight individuals using the thermal cycle sequence above and was separated using an automated sequencer (CEQ8000, SCIEX, Massachusetts, USA). Only three loci showed allelic polymorphisms, and these loci were used in the following experiments.
Preparation of DNA
Each sample was crushed in an individual 1.5 ml microcentrifuge tube, and total DNA was extracted using the DNeasyⓇ Blood and Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer’s experimental protocol. DNA from each sample was eluted into 200 μl of the elution buffer from the kit and preserved at 4°C until amplification by PCR. We used the three microsatellite loci mentioned above (My5, My6, and Mt5; Table 1) for genotyping.
Table 1: Pairwise Fst among the morphs in each sampling site. Statistical tests had been conducted by estimate 95% confidential limits of each value by 1000 boots strap resampling by locus. Asterisks mean p<0.01.
| EG | ER | JAG | JAR | JBG | JBR | |
| EG | ||||||
| ER | 0.0104 | |||||
| JAG | 0.0270* | 0.0610* | ||||
| JAR | 0.0143 | 0.0321* | 0.0324* | |||
| JBG | 0.0512* | 0.0680* | 0.0490* | 0.0249 | ||
| JBR | 0.0235* | 0.0386* | 0.0314* | 0.0049* | 0.0043 |
Genotyping
We amplified alleles at each locus using the following protocol. PCR reaction mixtures consisted of 0.5 μl of PCR buffer, 0.5 μl of dNTPs, 0.1 μl of each primer (10 pmol/μl, with one primer labeled by 0.025 μl of Taq polymerase (Ex Taq, TaKaRa, Osaka, Japan), 0.5 μl of DNA, and 3.3 μl of sterilized distilled water (total volume=5.0 μl). The following thermal cycle protocol was used: 2 min at 94°C, 28 cycles consisting of 30 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C followed by 7 min at 94°C. Then, the samples were kept at 4°C until electrophoresis. Alleles were separated using an automated sequencer with a 400 bp size marker (GenomeLab DNA Size Standard Kit-400, Beckman-Coulter, California, USA).
Sample categorizations and analyses for genetic structure among populations and morphs
Genetic differentiation between populations is measured by Fit, which can be partitioned into differentiation from population subdivision (Fst) and inbreeding within a subpopulation (Fis) [17]. “Hierarchical F analysis” using these indices with adequate categorizations of samples allows the degree of genetic differentiation among categories and estimates of its cause to be determined. For this analysis, we adopted the following sample categorizations. Since the main purpose of this study was to examine the degree of genetic differentiation between G and R, we treated each morph from a sampling shoot as a group. The second category was the sampling site (JA, JB, or E). Thus, we used six groups in this study, i.e., JA-G, JA-R, JB-G, JB-R, E-G, and E-R. First, we calculated Fst and Fis for the entire sample to estimate the degree of genetic differentiation from population subdivision (Fst) or inbreeding (Fis). Next, we calculated Fst among the sampled shoots within a sampling site and then among the sampling sites by morph. Fst was calculated for each morph between the sampled shoots and regressed on log-transformed distances (m) to test if there was any trend in geographical differentiation. To estimate whether G and R were differentiated over their whole genomes (i.e., for all the loci examined), we constructed a distance matrix based on the genetic distance among the site-morph groups (JA-G vs. JA-R etc.) and produced a Neighbor Joining (NJ) tree [18] from the obtained distance matrix. From the pattern of the identified clades, we estimated genetic relationships between the morphs. If both morphs are genetically differentiated at the whole genome level, R and G morphs should be found in separate clades. Furthermore, allelic frequency differentiations at each locus were examined statistically. Loci with highly differentiated allele frequencies may be linked with a locus that determines morph type even if there is no genetic differentiation between the morphs based on all loci.
Software used for genetic analyses
We used several software packages for genetic analysis of microsatellite data. Hierarchical F analyses were conducted using the “hierFstat” package in R (ver. 3.4.3). Genetic differentiation of allele frequencies of each locus was tested with the “Population Differentiation” option of Genepop (http://genepop.curtin.edu. au). Nei’s genetic distance was calculated with “gendist” in Phylip [19], and a NJ tree based on the obtained distance matrix was constructed with “NEIGHBOR” in Phylip.
Tables 2 and 3 shows Fst and Fis across several hierarchical levels. For the whole population (JA+JB+E), Fst was not significant between G and R, but Fis was significant at this level, meaning there was a degree of inbreeding. For the hierarchical level between the sites (JA-JB, E-JA, and E-JB), there was almost no genetic differentiation except for Fis for E-JA. Fst and Fis were not significant for the among shoots hierarchical level within each site. These results suggest that dispersal by winged forms should randomize genotypes within the spatial scale (approximately 800 m) of the study area.
Table 2: Pairwise comparisons of Fst between the 3 sampling sites. Statistical tests have been conducted by the same way in Table 1. Asterisks mean p<0.01.
| E | JA | JB | |
| E | |||
| JA | 0.0230* | ||
| JB | 0.0387* | 0.0165* |
Table 3: Hieralchycal F statistics in each morph and all individuals across 3 sampling sites. Statistical tests for Fit and Fis have conducted by GenePop on the web. Asterisks mean p<0.05.
| FIS | FST | FIT | |
| Green (3 populations) | 0.0891* | 0.0425 | 0.1278* |
| Red (3 populations) | 0.1075* | 0.0252 | 0.1300* |
| G+R (3 populations) | 0.1050* | 0.0261 | 0.1284* |
However, Figure 1 suggests that the geographic differentiation by distance was minimal because, for both the morphs, the intershot Fst significantly increased with the log-transformed distance between the examined shoots. The regression coefficients of both the morphs were significantly positive, although the slopes did not statistically differ between the morphs (Figure 1).
Figure 2 shows the NJ tree constructed from the distance matrix based on Nei’s genetic distance between G and R in each sampling site. The genetic distance was calculated across all loci. As shown by the tree, the distance between G and R from the same sampling site was smaller than that to other sites suggesting gene flow between the morphs. Thus, significant genetic differentiation across the whole genome level was not detected.
Figure 3 shows the frequencies of each allele at each locus in the whole population or by site. In the whole population, there was no significant difference in My6 and Mt5, but allele frequencies were highly significantly different between G and R at My5 (exact G test). When the data were analyzed by site, this trend did not change, although in some sites, there were minimal significant differences for My6 or Mt 5; however, in every site, the significance levels were much higher at My5.
A color polymorphism with green and red morphs is frequently observed in aphids [11,12,20], and many researchers have focused on maintenance mechanisms of this intraspecies polymorphism [9,12,21,22]. In Acyrthosiphon pisum, since ladybugs, a predator of aphids, prefer the red morph as prey and another predator (parasitic wasps) prefers the green morph, there is negative frequencydependent selection between the two morphs, and this mechanism balances the frequencies of both morphs [21]. However, since M. yomogicola is a myrmechophillus aphid, predation is rare because the attending ants repel most predators. Ants attending aphids obtain nutrients from their honeydew [23]. Myrmecophillous aphids or scale insects have been known to excrete honeydew with higher-quality sugars and amino acids in the presence of ants than without ant attendance [24]. For M. yomogicola, the main attending ant, Lasius japonicus, prefers G to R, i.e., the number of ants attending G is larger than that attending R [9]. This fact suggests that honeydew of G should be preferable to the ants. In contrast, in the absence of ants, the rate of increase of R was higher than that of G [9], which is consistent with a high cost of honeydew production for G.
The theory of natural selection [25] predicts that each shoot should be dominated by R but, surprisingly, attending ants manipulate the rate of increase of G to be similar to that of R [9], and thus, both morphs can coexist in ant attended colonies. The attending ants intentionally maintain R on the shoots that they inhabit because they manipulate the rate of increase of G only, meaning that they can discriminate between G and R. Thus, cheating by the R morph is not a possibility. Why does L. japonicus maintain R intentionally? There should be some other benefits to the ants for maintaining R. To date, this benefit has not been elucidated, but if there is no benefit to the ants, they should prey upon R producing poor honeydew. In fact, in a symbiosis with another aphid, L. japonicus (previously classified as L. niger) preyed upon aphid individuals that excreted lower amounts of honeydew [13]. Thus, the coexistence of G and R should be necessary for the ants to gain the maximum benefits from this symbiosis. Such a symbiotic system with multiple genetic morphs can evolve if there is a genetic trade-off between G and R in a beneficial trait.
Our results demonstrated the following: 1) Genetic differentiation by distance (Fst) is not significant within the entire study area (Table 1: maximum distance of approximately 700 m between sites), although significant positive regressions of Fst on log-transformed distances (m) were detected (Figure 1). The regression slopes for both morphs did not differ significantly, suggesting that the dispersion distance by the winged form did not differ between the two morphs. Fis was significantly positive within the entire study area and for several sampling sites (Table 1). Inbreeding is plausible because sexuparae of M. yomogicola are wingless and thus can mate with individuals on the same shoot. 2) The NJ tree showed that G and R from the same site were more closely related to each other than to any group in the other sites (Figure 2). Because Nei’s genetic distances were calculated for all three loci, this result suggests that intermorph mating occurred, meaning that G and R belong to a single gene pool. In fact, such mating has been observed in the field [26]. Although the inheritance mode of colors is unknown, body color must have some genetic basis because, in asexual reproduction, color is strictly inherited [12]. 3) In contrast to the above results, a locus (My5) showed highly significant differentiation in allele frequencies (Figure 3). In some areas, the other two loci had significant differentiation, but significance levels were always far lower than those at My5 (Figure 3).
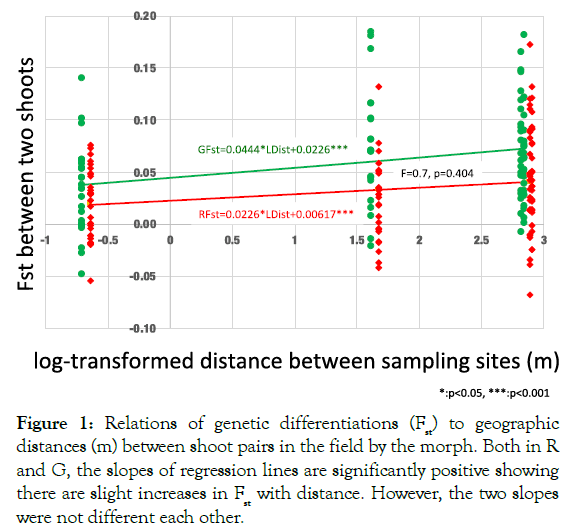
Figure 1: Relations of genetic differentiations (Fst) to geographic distances (m) between shoot pairs in the field by the morph. Both in R and G, the slopes of regression lines are significantly positive showing there are slight increases in Fst with distance. However, the two slopes were not different each other.
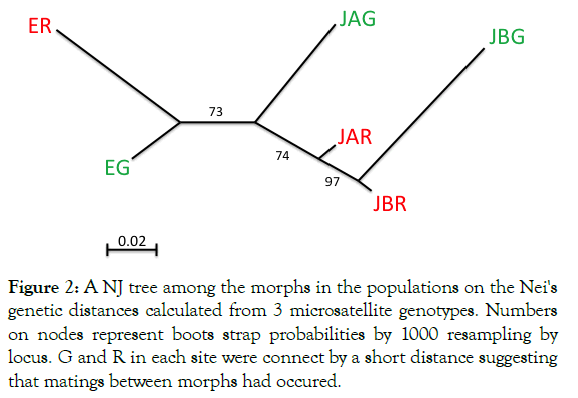
Figure 2: A NJ tree among the morphs in the populations on the Nei's genetic distances calculated from 3 microsatellite genotypes. Numbers on nodes represent boots strap probabilities by 1000 resampling by locus. G and R in each site were connect by a short distance suggesting that matings between morphs had occured.
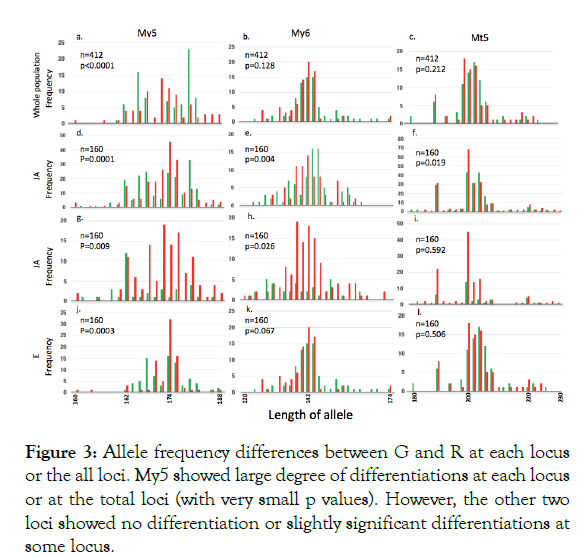
Figure 3: Allele frequency differences between G and R at each locus or the all loci. My5 showed large degree of differentiations at each locus or at the total loci (with very small p values). However, the other two loci showed no differentiation or slightly significant differentiations at some locus.
A reasonable explanation for these observations is My5 is linked to the locus (or loci) that determine the characteristic beneficial trait of each morph. When the whole genome sequence for both morphs and the primer sequence of My5 are identified, we can locate the position of My5 on the genome. Then, we can search for coding regions with a high degree of substitutions in amino acid sequences around My5. By comparing these protein sequences of multiple individuals of both morphs, we can specify the target gene because such gene(s) must be associated with two clades, each of which constitutes a single color morph, in a phylogenetic tree constructed from the DNA sequence of the gene.
There have been several studies on genetic differentiation between clones or populations of aphids [27-29]. These studies provide interesting perspectives on genetic polymorphisms under Darwin’s theory of natural selection (the maximization of the immediate reproductive rate of a lineage). However, in the symbiosis found in M. yomogicola, each partner appears to lose some of their immediate reproductive benefits to maintain the symbiotic system. L. japonicus may have reduced reproduction from maintaining the less nutritionally beneficial R morph. The potential reproductive rate advantage of R is suppressed by manipulation of the ants, and G has a decreased rate of increase potential from excreting high-quality honeydew. These losses of immediate benefits would contribute to the sustainability of these symbiotic relationships over time because, in a symbiotic system, every partner gains some fitness benefit from the system, and thus any cheating would result in loss of fitness for alternative honest behaviors. In other words, a symbiotic system requires biodiversity with genetic trade-offs among traits of each contributing member and simultaneously guarantees biodiversity within the system. By understanding the genetic basis of traits that can establish a symbiotic system, we may gain insight into the emergence and maintenance of the vast biodiversity in nature.
We thank to Dr. S. Watanabe for useful information of M. yomogicola
Citation: Hasegawa E, Murakami Y, Shiraiwa S, Kudo T (2019) Genetic Differentiation between Color Morphs of a Color-Polymorphic Aphid, Macrosiphoniella yomogicola. Entomol Ornithol Herpetol 8:217. doi: 10.35248/2171-0983.8.217
Received: 24-Apr-2019 Accepted: 31-May-2019 Published: 14-Jun-2019 , DOI: 10.35248/2161-0983.8.217
Copyright: © 2019 Hasegawa E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.