Journal of Research and Development
Open Access
ISSN: 2311-3278
ISSN: 2311-3278
Research Article - (2024)Volume 12, Issue 2
Autozygosity is the homozygous state of Identical-By-Descent (IBD) alleles, which can result from several phenomena. The increase in inbreeding leads to different negative effects such as a reduction in genetic variance, with a reduction in individual performance (inbreeding depression) and lower population viability. Nowadays, among several alternative methods to estimate inbreeding, F estimated from Run of Homozygosity (FROH) is considered the most powerful. This study aimed to provide a systematic review to comprehensive understanding of the structure and dynamics of genomic diversity and the impact of ROH studies over the last decade, highlighting directions and opportunities for future research. 406 publications were identified on WoS core collection and applied to bibliometric analysis available in the R environment. The analysis revealed an increased number of publications per year. China, Italy, and the United States are the countries that published the most on the topic, with the main contributions published in journals such as Animals and Frontiers in Genetics. This study highlights the growing interest in the functional analysis of ROHs driven by computational tools capable of characterizing these genomic regions. Noteworthy are the PLINK software and the detect RUNS package, utilizing a sliding window methodology for SNP analysis in the genome. Beyond a global crisis of genetic diversity loss, challenges encompass ecological issues, habitat fragmentation, and isolated populations, as well as the availability of genetic variation in species domesticated by humans, which forms the foundation of our food supply. A bibliometric analysis highlights the growing interest and progress of research in breeds of homozygosity and genetic diversity, driven by the advancement of genomic technology and international collaboration. Asia, America and Europe lead in knowledge production. Advances like PLINK and detect RUNS in R mean a quest for more accurate analysis. Emerging topics such as migration and correlation indicate an increasing focus on the impact of ROH standards. Challenges include refining analytical techniques and exploring regions of high heterozygosity, promising future research directions.
Genomic inbreeding; Run of homozygosity; Genetic diversity; Bibliometrics; Scientific production
The evolutionary history of a species can leave characteristics in the genome. Since the 1970s, molecular genetics has made rapid progress with the development of tools and technologies for manipulating DNA in vitro, which has allowed geneticists obtain the genomic sequences of model organisms. Contributing to the understanding of the chemical structure, functions and evolution of genetic material [1].
Since then, the genomes of different living beings have been sequenced with decreasing costs and improved technologies, such as Next-Generation Sequencing (NGS), Polymerase Chain Reaction (PCR) and advanced bioinformatics methods, allowing knowledge of the genomes of different species to be acquired and opening up possibilities for application in different areas of economy, health and species conservation in a way that is viable in terms of costs and routine [1-3].
In addition, the greatest challenge for modern genetics is not only sequencing genomes but transforming the data obtained available with precise and accurate information to improve the understanding of phylogenetic relationships and molecular evolution of the genomes of different taxa and to apply them as an innovative technology in studies of the diversity of life, with a view to application in medicine, genetic improvement, and genetic conservation [2,4].
Advances in whole-genome sequencing technologies and the use of high-density SNP arrays in scanning the genome for ROH has been proposed as an effective method for identifying IBD haplotypes are already available for humans and species of agricultural interest [5-7]. The scientific knowledge acquired has contributed to revealing unknown aspects of the genome and biological systems, resulting in a significant increase in research and publications in the field of molecular genetics [8].
In this context, significant advances in whole-genome scale approaches, when applied to genomic scans, can reveal genomic patterns of Runs of Homozygosity (ROH), highlighting the genetic variation that exists between individuals and enabling the detection of associations between genomic regions and adaptive phenotypes.
When the parents of an individual share a relatively recent common ancestor, they pass down large portions of their genomes that are identical by descent. Consequently, offspring increase homozygosity for that segment, thus creating a Run of Homozygosity (ROH) [9,10]. This phenomenon provides a valuable, often untapped resource for studying genetic diversity and the evolutionary history of various animal and plant groups [11,12].
Population evolution occurs due geographic effects, historical bottlenecks and isolations, along with cultural factors (anthropic effect), such as consanguineous mating, can result in elevated levels of homozygosity or autozygosity at both individual and population levels [11,13,14].
Longer regions of homozygosity increase the risk of recessive genes causing disabilities or fitness loss, reducing the viability of the organism and even the population [11,15,14]. Thus, understanding genetic variation within and between populations is relevant to determining the risk of population decline, as inbreeding is recognized as the main cause of adaptive reduction in different populations [13,15,16].
Therefore, it is important to characterize the genetic resources of different genomes to be used as support or reference in the formulation of guidelines for the conservation and sustainable reproduction of this biological diversity. Within this context, genomic analysis, with an emphasis on Regions of Homozygosity (ROH), can offer fundamental data on molecular evolution, adaptability and inbreeding coefficient based on regions of homozygosity (FROH) [15,17,18].
The main objective of this study was to gain an overview of the structure and dynamics of genetic diversity and run of homozygosity over the time, as well as to provide a visual summary of existing studies and to indicate directions and ideas for predicting the development of future studies. To this end, the Web of Science (WoS) was searched to identify publications that reveal the frontiers, and hotspots. And trends of research on genetic diversity and Runs of Homozygosity (ROH).
Bibliographic data recovery
The WoSs platform indexes more than 9,000 public and private academic institutions and organizes publications from more than 12,000 academic conferences, helping to make relevant decisions that enable them to shape the future of their institutions and research strategies [19].
On 21 February 2024, we conducted a bibliometric study of documents related to genetic diversity and runs of homozygosity published between 2009 and 2023 using the Web of Science Core Collection database (WoS). Here a sequence of keywords was adopted, including in order, the acronym TS= (genetic diversity AND runs of homozygosity). Articles and reviews were the main publication types. Meeting abstracts, editorial materials, and proceedings papers were excluded. Only publications type with ‘article’ were considered. Documents were uploading in BibTeX format and analyzed using Bibliometrix, R package. We analyzed the data frame using the Biblioshiny web app [9,20].
The number of publications over the years was subsequently measured to assess productivity, countries and regions, organizations, journals, keywords, and main references [9,20,21]. To minimize errors or lack of information and identify software used and animal species analyzed in each document, a manual review of the articles was carried out.
Country of the first authors
For each article, the address provided by the first author's research institution, including the geographical field of the countries included in the approach, was collected. When the first author's address was not identified, the second author's institute was [9,22].
Thematic evolution
The raw data were divided into different groups of consecutive years or periods to analyze the evolution of the field under study. The thematic evolution is organized in bibliometric maps and constructed by linking themes from one sub period with those from the next sub period through conceptual links. A communication path between themes is generated when there is a link between them, i.e., if they have common connective elements, such as keywords [9].
Publication quantitative
According to the data presented in the statistical graph of the research area, the scientific production of ROH and its genetic diversity vary over time. It began with modest production in 2009, followed by a slight increase in 2015, and as of 2017, there has been considerable growth in relevant studies in this field. It is important to note that the first approaches to genomic technologies applied to ROHs were recent, as demonstrated by the number of articles published in the first few years, as shown in Figure 1.
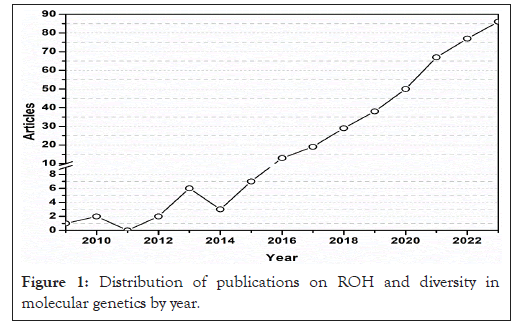
Figure 1: Distribution of publications on ROH and diversity in molecular genetics by year.
The increase in the number of publications from 2017 onward was probably due to the implementation and maturity of nucleic acid sequencing, as well as the high availability of information in public and private databases. Finally, between 2014 and 2022, the number of publications increased annually. Thus, as shown in Figure 1, studies of ROH and gene diversity have focused more on the field of molecular genetics in recent years. The main information collected for a review of the literature on Runs of Homozygosity (ROH) and genomic diversity is shown in Table 1. The results show that interest in this field is increasing, with an annual growth rate of 12.69%. The findings showed that ROH and genetic diversity are active and relevant fields, with each article receiving an average of 17.08 citations, demonstrating that ROH is important to the scientific community (Table 1).
| Description | Results |
|---|---|
| Timespan | 2009.2023 |
| Sources (journals) | 87.00 |
| Documents | 406.00 |
| Annual growth rate (%) | 12.69 |
| Document average age | 3.62 |
| Average citations per doc | 17.08 |
| References | 13.55 |
| Autor’s keywords (de) | 834.00 |
| Authors | 2317.00 |
| Authors of single-authored docs | 1 |
| Coauthors per doc | 7.64 |
| International coauthorships (%) | 51.23 |
Table 1: Main information on the survey carried out.
Most relevant journals associated with genomic diversity and ROH
All the relevant literature related to these fields of genetics was published in 87 sources or journals, 7 of which published more than 15 documents, Figure 2. In this respect, an analysis of the published documents reveals a significant interest in genetic diversity and ROH, as reflected in the approach of the journals.
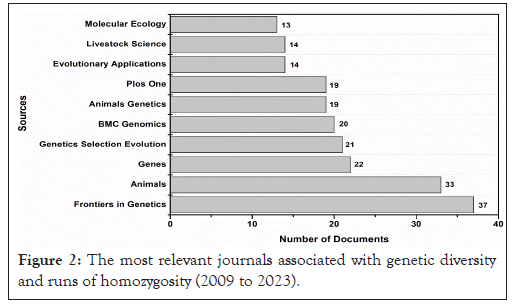
Figure 2: The most relevant journals associated with genetic diversity and runs of homozygosity (2009 to 2023).
International collaboration
In recent years, research into gene diversity and ROH has attracted interest among scientists worldwide. A bibliometric analysis revealed that researchers from 95 countries had significant academic output in the area, totaling 406 relevant publications in the period 2009-2023. Asian, American, and European research institutes have proven to be the main drivers of this scientific production. These studies show a global and multidisciplinary interest in this complex subject. In brief, ROH is a globally relevant and active area of study, with researchers from many different countries taking part. Table 2 provides an overview of the corresponding authors' countries (Figure 2).
| Country | No. of articles | Frequency of total | *SCP | *MCP |
|---|---|---|---|---|
| China | 79 | 0.302 | 56 | 23 |
| Italy | 40 | 0.153 | 28 | 12 |
| Estados unidos | 38 | 0.145 | 16 | 22 |
| Spain | 21 | 0.08 | 7 | 14 |
| Alemanha | 19 | 0.073 | 10 | 09 |
| Brazil | 14 | 0.053 | 08 | 06 |
| Canada | 14 | 0.053 | 08 | 06 |
| Iran | 14 | 0.053 | 03 | 11 |
| Netherlands | 13 | 0.05 | 03 | 10 |
| France | 10 | 0.038 | 04 | 06 |
| Total | 262 | 1.000 | 143 | 119 |
Note: *SCP=Single Country Publications; *MCP=Multiple Country Publications.
Table 2: The 10 most prolific countries in genetic diversity and ROH, corresponding author’s affiliation considered.
From the perspective of interconnectivity or networking the Single Country Publication (SCP), measures the level of intra-country collaboration; and the cross-country collaboration index, on the other hand, aims to monitor scientific collaboration between authors from different countries (Table 2).
China's intra-country cooperation is significantly more important than its intercountry cooperation (MCP). This phenomenon can be due to the scale and diversity of the sinology. In contrast, the U.S. is the third largest academic power, with 38 articles. However, it is worth noting that the proportion of interaction documents is greater in international searches (Table 2).
Table 3 presents the affiliations by field of study. The University of Palermo (Italy), Wageningen University and Research (The Netherlands), and the Institute of Animal Science (China) emerge as leaders with 36, 33, and 32 publications, respectively. The University of Edinburgh (United Kingdom) and Copenhagen University (Sino-Denmark) also make significant contributions, with 26 and 21 articles, respectively. The range of contributors continues remarkably, with associations from many countries, including United States, Spain and Brazil (Table 3).
| Affiliations | Number of articles | Articles (%) | Country |
|---|---|---|---|
| Univ palermo | 36 | 0.149 | Italy |
| Wageningen univ and res | 33 | 0.137 | Nettherlands |
| Inst anim sci | 32 | 0.133 | China |
| University edinburgh | 26 | 0.108 | USA |
| University Copenhagen | 21 | 0.087 | China |
| Cornel university | 19 | 0.079 | United State |
| Uniy calif los angeles | 19 | 0.079 | United State |
| Univ zagreb | 19 | 0.079 | Croatia |
| China agricultural university | 18 | 0.075 | China |
| Swedish univ agr sci | 18 | 0.075 | Sweden |
| Total | 241 | 1.00 | - |
Table 3: Top 10 affiliations in the publications involving genetic diversity and ROH.
Most relevant document
The 10 most cited articles had a total of 1,858 citations (Figure 3). These articles were distributed across 9 different scientific journals. The most cited article is the paper titled " Genomic Patterns of Homozygosity in Worldwide Human Populations", written and published in The American Journal of Human Genetics (SJR factor 9,8), with a total of 314 citations (Figure 3) [15].
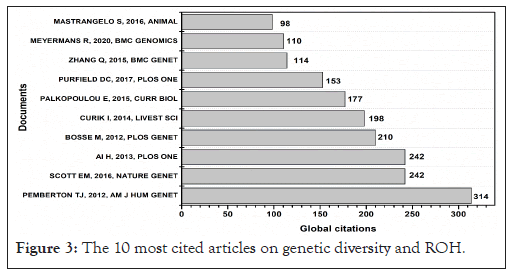
Figure 3: The 10 most cited articles on genetic diversity and ROH.
Although the journal nature genetics is not the main resource, it is important to note that this article received a significant number of citations. Interestingly, the authors of the most cited articles include two (the authors) of the 10 most productive researchers.
Notably, eight of the ten most cited articles were published after 2013, confirming the results observed in annual scientific production (Figure 1). The growing trend of research into genetic polymorphisms and ROH, especially since 2017, demonstrates the heterogeneity of population genomics studies and the detection of genetic hybridization through ROH.
Researchers are exploring new approaches, such as how evolutionary factors, such as demography, assembly, and specific selection pressures, affect the population’s genomic structure. This study reports on a comprehensive population genome analysis.
Most frequent keywords
From a quantitative analysis, 3121 keywords were identified that were used repeatedly by the authors of the publications under analysis. These terms, endowed with specificity, effectively represent the content described in the texts. When evaluating the main meaning of these words, "homozygosity", "run" and "genetic diversity" were distinguished. These terms are emerging as central pillars of gene diversity and ROH research. These concepts have evolved remarkably over the years, revealing progressive interest and deepening.
Over time, the information demonstrated a continuous and significant increase in keywords. Figure 4. In 2023, as an example, the word "homozygosity" is recorded a significant total of 154 times, while "breed" reaches the mark of 143 occurrences (Figure 4).
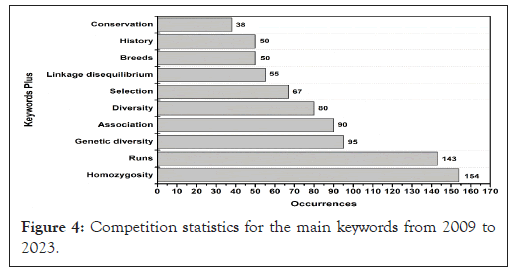
Figure 4: Competition statistics for the main keywords from 2009 to 2023.
In addition, the term "genetic diversity" appeared 95 times, showing a strong interest on ROH to elucidate biodiversity availability. The analysis also revealed that 'association' or ‘selection', has a relevant approach, with occurrences of 90, and 67 records, respectively. Also notable is the frequency of “linkage disequilibrium" term appears. In addition to the significant presence of goal of "conservation” as well as the importance of "history", which signals an increase in its occurrence over the years, see (Figure 4).
Thematic map
The use of a thematic map becomes relevant when co-word analysis is used to trace the structure of a specific discipline. The main objective of this approach is to trace the conceptual structure using a term co-occurrence network to map and group keyword terms, subheadings, or abstracts in a bibliographic compilation [20,23].
Data analysis under a quantitative approach revealed the emergence of four different thematic regions, where each region was in a specific quadrant (Figure 5). In the upper right quadrant, specialization themes such as "homozygosity", "runs", "genetic diversity", "association" and "linkage" are highlighted (Figure 5).
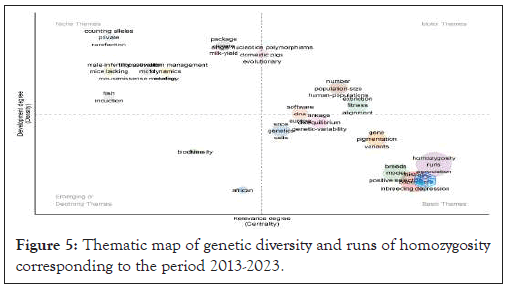
Figure 5: Thematic map of genetic diversity and runs of homozygosity corresponding to the period 2013-2023.
These themes stand out due to their high density and outstanding centrality and play a fundamental role in structuring research fields, which has become a relevant component [9,20]. Conversely, the topics in the upper left quadrant preserve intrinsic links and correspond to areas of rapid development, which is reflected in their greater density, despite their more modest centrality. These topics, such as "r packhage", "demografic history", "hybridization" and "models", are specialized and peripheral (Figure 4).
In the bottom left corner of the map, topics considered marginalized and underdeveloped can be examined; although these topics are sound to a certain extent, their centrality and decreasing density indicate incipient growth or even the possibility of expansion. There are links to topics such as "genomes" and "positive selection" (Figure 5).
Finally, the topics in the lower right quadrant are substantial, wide-ranging, and cross-cutting, characterized by their high centrality. However, they have a relatively low density. Topics such as "homozygosity", "evolution", "inbreeding depression", "mutations", "sequence" and "alignment" in this quadrant signal convergence in the study of genetic diversity and ROH in many species, from humans to different pets. However, these topics are still under development and will form the basis and coverage for the near future.
Applied methodologies
The functional analysis of Runs of Homozygosity (ROH) is supported by computational tools that play a key role in detecting and characterizing these genomic regions. Among the usual tools used to analyze SNP microarray data, PLINK software the most used program [24,25]. A total of 37 articles published between 2013 and 2017 demonstrated the efficacy of utilizing PLINK software, with its superiority observed in 33 of these studies (Figure 6).
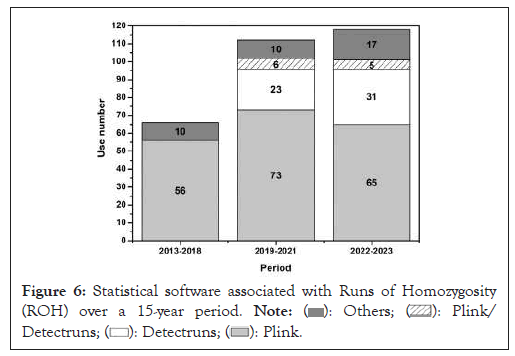
Figure 6: Statistical software associated with Runs of Homozygosity
(ROH) over a 15-year period. 

However, relevant transformations have occurred in the software applications landscape. From 2018 to 2020, the PLINK software remained the most used, being cited in 60 studies. On the other hand, the rise of the R detect RUNS program was seen as a viable option, with its use in 12 studies during this period.
Between 2021 and 2023, there was a significant increase in the variety of software used. PLINK continued to be the most used, present in 86 searches, followed by detect RUNS 0.9.5, which appeared in 48 articles. Furthermore, other software programs were explored, highlighting the search for more comprehensive and accurate analytical methods during this period.
Publications analysis based on animal diversity
The analysis of approximately 275 documents allows us to verify that ROH research requires a careful sampling methodology to assess the distribution of genotypes in animal populations, both domestic and wild. Based on the data provided, a sample covered a diverse set of species, including Human (25), cattle (68), sheep (54), pig (35), goat (23), chickens (25), other domestic and wild animals (59) (Figure 7).
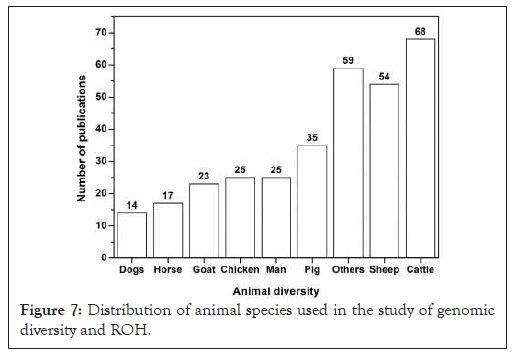
Figure 7: Distribution of animal species used in the study of genomic diversity and ROH.
Quantitative publications per year
Quantifying genetic diversity and levels of kinship in humans, plants, and animals is a key element for evolutionary biologists and agricultural scientists, enabling the development of new methods aimed at revealing knowledge applied to genetic improvement and the conservation of populations at risk of extinction [19,26]. It was revealed by an increase in the number of publications. This increase may be directly related to the growing availability of biological information which makes the application of genomic technologies more accessible. This has allowed these technologies to be adopted with relatively less investment in research and facilitated the creation of larger laboratories [9].
This development represents a significant milestone, as the significant improvement in the availability of genomic sequences enables the creation of genetic maps for animals of zoo technical interest. These advances have been documented by several experienced authors, who have exploited the potential of the data to improve animal management, genetic conservation, and selection [11,17,26,27].
Thus, broadening access to genome sequencing data not only expands the frontiers of scientific knowledge but also encourages the development of more effective management strategies, benefiting sectors such as livestock production. This field continues to play a key role in promoting sustainability and preserving crops, thus enriching the field of evolutionary and agricultural research [17, 27,28].
Main countries and institutions linked to publications involving ROHs
The Republic of China ranks first in the number of publications but most of the publications are from a single country (Single Country Publication-SCP) (Table 2) and (Figure 2). Multiple Country Publication-MCP), represents more than half of all publications, and the U.S. ranks first with multiple contributions, suggesting that the results are associated with the availability of qualified human resources mixed with technological capacity and public and private investment in scientific research.
It should also be noted that China, Italy, the United States stand out as key collaborating countries in the development and dissemination of genomic technologies, genetic studies, and bioinformatics related to polymorphisms, ROH, and inbreeding [4, 29-32]. Brazil in South America and Iran in Asia are recognized as emerging countries in this field [33,34] (Tables 1 and 2).
They have a strong interest in population genetics, with a focus on human and animal genomic inbreeding and a particular emphasis on recessive diseases, animal husbandry, and the conservation of endangered species. the US focuses on human and nondomestic animal populations, while China and most other countries concentrate their studies on breeding and diversity of domestic animals.
China, USA, and Italy have played significant roles in scientific research related to animal production. More developed countries with higher incomes typically possess greater resources for scientific research, which elucidates the prominence of these nations in the analyses. China's economic importance is also noteworthy, with contributions to the development of technologies and investigative efforts applied to agriculture and livestock [35-37].
Strategic areas and thematic maps
The top right quadrant of Figure 5 contains topics related to population size, human population, and extinction. The relevant principles and concepts related to these topics are described in detail in the "population structure", "congenital recessive disease" and "gene conservation". Most of the concepts and principles reflect the study of the molecular basis of gene diversity and ROH in human, domesticated, and wild populations [12,16,17].
The matching and software topics in this quadrant often include related concepts such as genomics and genetics. These issues stand out because they are associated with nucleic acid sequencing technologies that have organized and still generate large amounts of data that allow comparative studies of different biological species and populations around the world using bioinformatics tools [1,38,39].
In the top left quadrant of Figure 5, there is a specific topic and associated field density. This quadrant highlights the topics of allele counts, privacy, dispersal, and packing, suggesting links between this topic and genomic identification, species characterization, and population decline.
Finally, owing to these thematic maps, scientists can map the evolution of research into runs homozygosity and gene diversity, identifying areas of specialization, interconnections between topics, and the relative relevance of each key term. This bibliometric analysis allows researchers to understand the panorama of knowledge in this area and to identify research gaps, new directions, and possible interdisciplinary collaborations [22,25].
Finally, thematic maps provide a panoramic view of the controversies surrounding racial similarity and genetic diversity. These visual resources highlight both the surrounding areas and the main elements of the research, revealing the complexity of the relationships between the subjects. Understanding these dynamics is important for the development of this field and the promotion of important discoveries in genetics and evolution.
Software and statistical approaches
The conversation about software tools used to analyze Runs of Homozygosity (ROH) and regions rich in heterozygosity is comprehensive and essential in current genomic research. In this study, a quantitative assessment was made of the use of programs capable of identifying and characterizing ROH in the genome. The results make it possible to visualize different programs. PLINK software that uses a Sliding Window (SW) to manage and analyze data on individual genotypes has a high frequency of use [26,40, 41].
Additionally, the Detect RUNS, a well-known tool, is an R package created to identify both ROH and stretches of heterozygosity. This software uses consecutive-windows and sliding-windows approaches, offering features to summarize findings and calculate inbreeding using ROH data [17,40].
Besides detecting runs, the package offers several convenient functions to summarize and plot results, and to compute inbreeding based on ROH (runs of homozygosity). Runs of Heterozygosity. Heterozygosity-rich regions, are a relatively novel concept, introduced [42]. One of the objectives of this project is to develop methods and code, and to explore parameters, for the detection of “runs of heterozygosity” in diploid genomes (namely, animals).
In addition to the sliding windows and consecutive run methods employed by detect runs, there exist other software and methodologies suitable for detecting ROH.
Consecutive run method
In Marras et al., starting from the first SNP, if this is homozygous it is considered to be potentially in a ROH [15]. The next SNP is evaluated: If heterozygous, the process stops here; if homozygous, a potential ROH may be detected. This goes on until the minimum n. of SNP in a ROH is met: A ROH is thus detected and will span until a heterozygous SNP is encountered.
No overlapping windows are used in this method. Parameters based on heterozygous and/or missing SNP loci can be tuned to relax the identification of ROH. This method can be adapted to the detection of “Runs of Heterozygosity” (ROHet) by simply inverting the detection of homozygous loci to that of heterozygous loci (and adjusting parameters). Python code from the Zanardi software package, was expanded and translated into R for the implementation of the consecutive method in detect RUNS.
An alternative method it is the sliding windows
The software Plink Purcell et al. implements a sliding-window algorithm to detect runs of homozygosity in diploid genomes (specifically the human genome) [40]. This is based on the following [43]:
• From the first SNP, a sliding window of n SNPs is used to scan the genome (chromosome by chromosome).
• When a stretch of m homozygous SNP (m ≤ n) is identified, the n. of sliding windows covering such stretch is counted at each SNP locus.
• The n. counted at 2) is divided by the total n. of windows covering each locus (increasing from 1 to n, initially, then constant at n, eventually decreasing from n to 1).
• SNP loci for which the ratio at 3) is ≥ 5% are classified as being in a ROH.
• At 2), flexibility parameters may be applied: n. of heterozygous SNP loci allowed in a ROH, n. of missing SNP loci.
This procedure was translated into R code and adapted also to the detection of “Runs of Heterozygosity” (ROHet), The trick should reside simply in counting heterozygous instead of homozygous SNP stretches at point: the minimum number of SNPs included in the HRR; the number of missing or opposite genotypes; the maximum gap between consecutive SNPs; the minimum HRR length. The R code to implement this algorithm was developed in detect RUNS.
R was mainly used to write functions in detect RUNS. The R package format was chosen to facilitate documentation, organization of the code and its distribution and reproducibility [41,44]. The R package detect RUNS has seen a significant surge in its utilization and is likely one of the most swidely adopted programs for analyzing SNP genotypes and Runs of Homozygosity (ROH) and Heterozygosity-Rich Regions (HRR), across both animal populations [45-48].
Bibliometric analysis has demonstrated considerable progress in producing studies related to Runs of Homozygosity (ROH) and genetic diversity over the years. Initially discreet in 2009, there was a significant increase from 2017 onwards, showing growth in this field, driven by the advancement of genomic technologies and the greater availability of information in public and private databases. Many relevant articles were identified across 87 sources or journals, demonstrating the increasing interest in genetic diversity and Regions of Homozygosity (ROHs). Furthermore, the bibliometric analysis highlighted considerable international cooperation in this field, with researchers from 95 countries contributing to academic research. Study centers in Asia, America and Europe stood out as the main drivers of scientific production in this area.
The research also highlighted advancements in the methods and technology employed in the study of ROHs. The PLINK software was commonly utilized for the examination of SNP microarray data, demonstrating its efficacy in detecting and describing homozygous genomic regions. Furthermore, the emergence of the detect RUNS tool in R as a potential alternative indicates a quest for more thorough and precise analytical approaches. The analysis of keywords unveiled new central topics like "homozygosity", "migration", "genetic variability", "correlation" and "ancestry", showing an increasing depth and curiosity in exploring the patterns of ROHs and how they influence genetic diversity across generations. These themes also demonstrate a variety of perspectives, ranging from research on human communities to both domestic and wild fauna.
The steady advancement of research into ROH and genetic diversity indicates a continued and important interest in this field. However, we face challenges, such as the need to improve more advanced analytical techniques to deal with complex and diverse data sets. Future investigations should delve into new areas, such as regions rich in heterozygosity that are capable of detect differences between chromosomal regions, providing a more detailed picture of genetic diversity.
The foundation of support to research of Piauí (FAPEPI).
Conceptualization, F.A.D.S., R.C.C., J.O.M., M.M.B., L.C.B.C., F.B.B. and A.M.A.; Methodology, F.A.D.S., R.C.C and A.M.A.; Validation, F.A.S., R.C.C., J.B.C., F.B.B. and A.M.A.; Formal analysis, F.A.D.S., R.C.C and A.M.A.; Investigation, Resources, F.A.D.S.; and A.M.A; Writing-original draft, F.A.D.S; Writingreview & editing, F.A.D.S., R.C.C and A.M.A; Visualization, F.A.D.S., R.C.C and A.M.A; Supervision, F.A.D.S., R.C.C and A.M.A; All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
[Crossref] [Google Scholar]
Citation: Sobrinhoa FDAD, Coelho RC, Mourab JDO, Baja MM, Carvalho LCB, Brittoa FB, et al. (2024) Genetic Diversity and Runs of Homozygosity (ROH): A Portrait of the Quantitative Academic Publication Dynamic and Scientific Metadata. J Res Dev.12:256.
Received: 24-Apr-2024, Manuscript No. JRD-24-30928; Editor assigned: 29-Apr-2024, Pre QC No. JRD-24-30928 (PQ); Reviewed: 14-May-2024, QC No. JRD-24-30928; Revised: 21-May-2024, Manuscript No. JRD-24-30928 (R); Published: 28-May-2024 , DOI: 10.35248/2311-3278.24.12.256
Copyright: © 2024 Sobrinhoa FDAD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.