Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Research - (2022)Volume 10, Issue 12
Background: In Ethiopia, an estimated 68 million people are at risk of malaria–60% caused by Plasmodium falciparum and 40% by Plasmodium vivax. The national elimination program has begun since 2016 with a vision of malaria-free country by 2030. The radical cure of P. Vivax with the drug primaquine is an important component of the elimination strategy. However, Primaquine causes acute hemolytic anemia in glucose-6 phosphate dehydrogenase enzyme deficient patients and is a threat to P. vivax elimination. G6PD is a cytoplasmic enzyme for all human cells that is involved in the pentose phosphate pathway of metabolic reaction and protects red blood cells from cellular oxidative damage by detoxifying free radicals. This study is therefore carried out to determine the G6PD deficiency prevalence among malaria-suspected patients in the study sites.
Methods: Health facility-based cross-sectional study was conducted in 2021 at Shele and Lante health centers in Southern nations and nationalities people region of Ethiopia. A total of 858 self-presented malaria-suspected patients were enrolled in this study. The socio-demographic and clinical information of the study participants were collected using a pre-validated questionnaire, entered into Epi Info™ 7 software, and analyzed using SPSS V.20 statistical software. Finger prick blood samples were collected for onsite Care START G6PD biosensor analyzer test, malaria smear microscopy, and Dried Blood Spot (DBS). The DBS samples were used for molecular confirmation of G6PD deficiency.
Results: From a total of 858 study participants enrolled in the study 49.3% (423) were males with the median and interquartile age range of 26 and 21 years, respectively. Of all the study participants, 14.3%, 9.3%, and 4.1% were smear-positive by microscopy for P. falciparum, P. vivax, and mixed parasites, respectively. The phenotypic Care START biosensor analyzer G6PD deficiency rate was 4.8% (41/858) while the molecular genotyping results analyzed in selected 13 patients have shown G6PD gene mutation in 10 (76.9%) of the samples. Particularly G267+119C/T mutations were seen in 9 of 13 (69.2%) while A376G, and G1116A were seen in 3/13 (23.1%) participants equally. In addition, new mutations such as A376T (A→T) in 2/13 and G1116T (G→T) in 1/13 of participants were also identified.
Conclusion: The result implied that G6PD deficiency among the study participants is not significantly high. In addition, the G267+119C/T mutation was the most frequent variant reported in this study. Therefore, it is recommended to consider hemolysis risk while prescribing the primaquine drug in the study area.
G6PDd; Hemoglobin; Plasmodium vivax; Plasmodium falciparum; Primaquine
Malaria is one of the most important public health problems where more than 68 million peoples of Ethiopians are at risk. In Ethiopia, the major etiologic agent of malaria is caused by P. falciparum accounting for about 60% followed by 40% by P. vivax [1]. Even though the complete epidemiologic distribution of P. malarae and P. ovale is not well studied, few cases (<1%) have been reported from some parts of the country [2].
A key malaria elimination strategy indicated in the national malaria elimination roadmap is case management through accurate diagnosis and prompt of treatment using the safe anti-malaria drug [3]. As part of this, the Minister of Health (MoH) has endorsed primaquine use for the treatment of liver stage (hypnozoite) parasites for the radical cure of P. vivax [4,5]. However, primaquine drugs cause severe hemolysis in individuals with Glucose-6 Phosphate Dehydrogenase (G6PD) enzyme deficiency [6,7].
The Glucose-6 Phosphate Dehydrogenase (G6PD) enzyme is a cytoplasmic enzyme in the pentose phosphate pathway of all human cells in the body preventing cellular oxidative damage to red blood cells by detoxifying free radicals [8-10].
The G6PD Deficiency (G6PDD) is an X-linked genetic disorder caused by the coding gene's mutation resulting in free radical-mediated oxidative damage to red blood cells and causing acute hemolysis. It is the most common enzymatic disorder of red blood cells affecting approximately 400 million people worldwide [11] with the highest genetic disorder in malaria-endemic areas of Africa, Asia, the middle east, Latin America, the Caribbean, and Mediterranean countries [12-16].
Although a comprehensive national study has not been conducted, the prevalence rate of G6PD deficiency in Ethiopia is estimated at about 1-3% [17] with wide range of variation in lowland areas of the country ranging from 1.4% to 14% [13-18].
Most of the time, G6PD deficient individuals appear normal and clinically silent until exposed to free-radical elements. This is more common in hemizygous males than females since females can be either homozygous or heterozygous. Those individuals who had RBC’s G6PD enzyme activity <30% of the normal G6PD activities had hemolysis in common after administration of primaquine [19-21] which becomes higher in P. vivax infected patients because of a higher dose of primaquine (0.25–0.5 mg for 14 days) than in P. falciparum (a single dose of 0.25 mg) [4,21-23].
Based on the biochemical and physicochemical characteristics of G6PD, more than 400 variants have been described to date, of which approximately 186 variants are related to G6PD deficiency [24]. Most of which are single nucleotide substitutions [9]. In Africa, the common genotyped variants were G6PD B+(wild type), G6PD A+(376), G6PD A-(202), and Mediterranean variant (563) [1,10,15,25]. Of these variants, A+ is a non-deficient G6PD-type variant, while A- and Mediterranean variants are known deficient variants. In Ethiopia, studies have estimated the A-(G202A) and A+(A376G) variants prevalence was 3.5%, and 17%, respectively [1,26]. Therefore, this study aimed to determine the current prevalence of G6PD deficiency and its genetic variants among malaria suspected patients in Shele and Lante health center, Southern Ethiopia.
This cross-sectional study was conducted on 858 malaria suspected consented patients, who had the signs and symptoms of malaria and self-presented at Shele and Lante health center of Arba-Minch Zuria woreda, Southern Ethiopia from February 2021 to April 2021. Each study participant was subjected to finger prick blood collection for blood film preparation, and malaria Rapid Diagnostic Tests (RDT). In addition, venous blood was collected using EDTA tubes for a quantitative rapid CareSTART G6PD biosensor analyzer test (sensitivity=92-100%, and specificity=92-94% against the spectrophotometric method) following the procedure described by Bancone et al. [27]. The sub-set of samples that have shown G6PD deficiency by G6PD biosensor analyzer tests were further subjected to molecular genotyping of G6PD variant mutations. The genotyping test was conducted on randomly selected 13 samples (10 G6PD biosensor analyzer deficient samples (6 male and 4 female) and 3 female intermediate samples) following a standard procedure [1].
Briefly, four PCR assays were conducted to determine the G6PD gene mutations of exons [3-11]. PCR used a 20 μl reaction mixture containing 2 μl of genomic DNA (~50 ng/μl), 10 μl of 2 × Maxima SYBR Green PCR Master Mix (Thermo Fisher), and 0.3 uM of each forward and reverse primer. For each PCR assay, water was used in a separate reaction as a negative control. Amplifications were done through an initial denaturation at 94°C for 3 minutes, followed by 38 cycles at 94°C for 30 sec, 58°C for 30 sec and 72°C for 60 sec, with a final 6 min extension at 72°C. Then, the PCR products were run by gel electrophoresis with 1.5% agarose gel at 120 volts for two hours and sequenced on an ABI 3730xl DNA analyzer following standard protocols (Genewiz Inc., La Jolla, CA). Sequences were analyzed using BioEdit. All sequences were aligned to the NCBI reference sequence to check the specificity of the PCR products. Amplified PCR products with poor sequencing quality or exhibited singleton mutations were repeated [1].
The patients who were malaria positive either by microscopy and/or RDTs (Care START Pf/Pv (HRP2/PLDH; Lot No MV19861)) were treated with antimalarial drugs following the national treatment guideline. The Care START G6PD biosensor analyzer test results were interpreted as described in Table 1 below. Demographic and laboratory analysis data of the study participants were captured using a pre-validated questionnaire, entered into Epi Info™ 7 software and analyzed using SPSS V.20 statistical software. Descriptive statistics were used to describe the frequency of each variable. A binary logistic regression model and independent t-test were performed to assess factors associated with G6PD deficiency and genetic variants among study participants. The phenotypic classifications of the G6PD enzyme were performed based on the WHO classification by calculating the Adjusted Male Median (AMM). P-value <0.05 was considered statistically significant.
| Gender | Type | G6PD range | Calculated G6PD_Hemoglobin ratio |
|---|---|---|---|
| Male | Deficient | <30% of the AMM | <1.8 |
| Normal | >30% of the AMM | >1.8 | |
| Female | Deficient | <30% of the AMM | <1.8 |
| Intermediate | 30-80% of the AMM | 1.8-4.8 | |
| Normal | >80% of the AMM | >4.8 |
Note: AAM=Adjusted Male Median.
Table 1: Classification of G6PD enzyme activities among males and females.
Socio-demographic characterstics and malaria diagnostic results using CareStart Pf/Pv RDT and Microscopy
A total of 858 malaria suspected patients, 49.3% (423) male, the median age of 26 years and interquartile range of 21 years were enrolled in the study. All study participants had fever (100%) followed by 93.9% headache, 53.3% joint and muscle pain, 35.2% fatigue, and 18.3% abdominal discomfort. Patients who had headaches showed a significant association with malaria positivity (P<0.05) (Table 2).
| Socio-demographic characteristics N=858 | N (%) | Microscopic positivity N (%) | P- value | G6PD Deficient (N%) | P-value | |
|---|---|---|---|---|---|---|
| Sex | Females | 435 (50.7) | 103 (12.1) | 23 (2.7) | ||
| Males | 423 (49.3) | 136 (15.9) | 18 (2.1) | 0.7 | ||
| Age group | >15 years old | 690 (690) | 174 (20.4) | 37 (4.3) | ||
| <5 years old | 15 (1.7) | 9 (1.1) | 0 | |||
| 5-14 years old | 153 (17.8) | 56 (6.6) | 4 (0.5) | 0.07 | ||
| Place of residence | Urban | 389 (45.3) | 85 (10) | 5 (0.6) | ||
| Rural | 466 (54.3) | 153 (18) | 33 (3.9) | 0.002 | ||
| History of malaria infection | No | 172 (20.3) | 36 (4.2) | 5 (0.6) | ||
| Yes | 683 (79.6) | 202 (23.8) | 33 (3.9) | 0.7 | ||
| P. falciparum Positivity | Negative | 695 (81.5) | NA | 24 (2.8) | ||
| P. falciparum | 158 (18.5) | NA | NA | 17 (2.0) | 0.6 | |
| Malaria positivity | Negative | 614 (72.0) | NA | 19 (2.2) | ||
| Positive | 239 (28.0) | NA | NA | 22 (2.6) | 0.3 | |
| P. vivax positivity | Negative | 738 (86.5) | NA | 32 (3.8) | ||
| P. vivax | 115 (13.5) | NA | NA | 9 (1.1) | 0.9 | |
| Clinical signs and symptoms | ||||||
| Headache N=858 | No | 52 (6.1) | 5 (0.6) | 1 (0.1) | ||
| Yes | 806 (93.9) | 234 (27.4) | 0.007 | 40 (4.7) | NA | |
| Fatigue N=858 | No | 556 (64.8) | 162 (19) | 27 (3.1) | ||
| Yes | 302 (35.2) | 77 (9) | 0.9 | 14 (1.6) | NA | |
| Muscle and Joint pain N=856 | No | 399 (46.5) | 119 (14) | 13 (1.5) | ||
| Yes | 457 (53.3) | 120 (14.1) | 0.99 | 28 (3.3) | NA | |
| Shaking and chills N=858 | No | 457 (55.4) | 128 (15) | 35 (4.1) | ||
| Yes | 383 (44.6) | 111 (13) | 0.15 | 6 (0.7) | NA | |
| Sweating N=858 | No | 533 (62.1) | 148 (17.4) | 29 (3.4) | ||
| Yes | 325 (37.9) | 91 (10.7) | 0.85 | 12 (1.4) | NA | |
| Anorexia N=858 | No | 593 (69.1) | 129 (15.1) | 22 (2.6) | ||
| Yes | 265 (30.9) | 110 (12.9) | 0 | 19 (2.2) | NA | |
| Abdominal discomfort N=858 | No | 701 (81.7) | 210 (24.6) | 38 (4.4) | ||
| Yes | 157 (18.3) | 29 (3.4) | 0.003 | 3 (0.3) | NA | |
Note: NA=Not applicable
Table 2: Sociodemographic characteristics and their associated factors for G6PD deficiency among malaria suspected patients.
P. falciparum was detected in 22.1% of participants with RDT, 22.4% with laboratory personnel microscopy, and 14.3% with expert microscopists' while the performance of RDT, laboratory personnel microscopy, expert microscopists' for P. vivax was 11.7%, 11.3%, and 9.3%, respectively even though the difference is not statistically significant (P>0.05) (Figures 1 and 2).
Figure 1: Location of Shele and Lante Health Center, in Arba Minch
Zuria Woreda, South Nation Nationalities and Peoples Region,
Southern Ethiopia. 
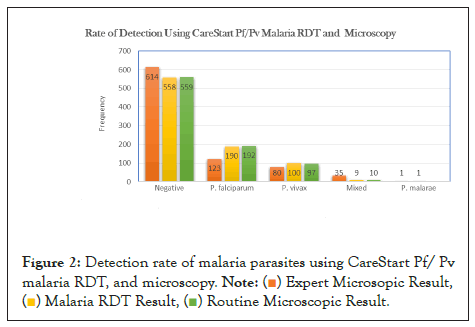
Figure 2: Detection rate of malaria parasites using CareStart Pf/ Pv
malaria RDT, and microscopy. 
G6PD deficiency and associated factors among study participants
The G6PD biosensor analyzer median and interquartile range of the G6PD enzyme level was 5.9, and 4.2, respectively where 4.8% of the study participants were G6PD deficient with less than 1.8 enzyme activity. Males accounted for about 2.1% [18] while G6PD deficient and intermediate females accounted for about 2.7% [23] and 29% (126) respectively.
About 89.5% of G6PD deficient were above 14 years old. However, no significant association was observed with G6PD deficiency despite the difference in frequency. Based on the expert microscopic result, of 41 G6PD deficient patients, 19 were negative while 22 (P. falciparum=13, P. vivax=5, mixed=4) were positive for malaria parasites. There were no significant association between P. falciparum infection and G6PD deficiency although higher chance of being deficient than non-P.falciparum participants was expected (AOR=1.5; P=0.6). Similar association were observed in all malaria-positive cases (AOR=2.6; P=0.3) and P. vivax positive cases (P>0.05). G6PD deficiency was four times more likely to occur in a rural area than compared to the urban territory (P=0.002) (Table 2).
Among G6PD deficient patients, only two of them had shown severe anemia exhibiting below 5 g/ dl of hemoglobin, of which one was pregnant and P. falciparum positive with high parasitemia (>2000 parasite/ µl of WBCs) while the other was negative for malaria parasites.
Of the study participants, 27.9% (239) were positive for malaria parasites of which 26.7% had a corresponding parasite load count. Then, the corresponding parasite load of positive cases was compared between G6PD deficient and normal. The correlation coefficient between the G6PD enzyme level and the parasite load was 0.08 (P=0.2), the correlation was statistically not significant. This was also confirmed by logistics regression output (P=0.5, 95% CI, 0.132-2.822). However, the overall parasite load proportion of G6PD normal versus G6PD deficient had a statistically significant difference (P<0.05) (STATA V.14).
Genotype characteristics of glucose-6 phosphate dehydrogenase
Of a subset of 13 patients, 10 (76.9%) had shown mutation in their G6PD gene while 3 (23.1%) hadn’t shown mutation. Among patients who had no mutation, none of them were phenotypically intermediate for G6PD. Of ten (10) participants who had mutations (7 G6PD deficient and 3 intermediate deficiency of G6PD), five of them had shown genotypic polymorphism of whom five had mutation due to single nucleotide substitution. Among participants with polymorphic mutation, one study participant had a triple mutation at three nucleotide positions (A376T, G267+119C, and G1116A) while the remaining four had double mutations at their G6PD gene.
G6PD A+ (A → G and A → T) at nucleotide position of 376 was detected in 3 (23.1%) of the study participants. Of these, A → T was shown in 2 (15.4%) of the phenotypically deficient, female participants. G267+119C/T and G1116A were also detected in nine (69.2%) and three (23.1%) of the study participants, respectively. No mutation was detected for G6PD A-(G202A) and Mediterranean type (C563T) among all participants (Table 3).
| Target genes | Frequency (%) | ||
|---|---|---|---|
| N=13 | |||
| A376G | N (%) | 10 (76.9) | |
| Wild Type (%) | 7 (53.8) | ||
| Mutant | A→G (%) | 1 (7.7) | |
| A→T (%) | 2 (15.4) | ||
| G202A | N (%) | 9 (69.2) | |
| Wild Type (%) | 9 (69.2) | ||
| Mutant | G→A (%) | 0 | |
| G267+119C/T | N (%) | 11 (84.6) | |
| Wild Type (%) | 2 (15.4) | ||
| Mutant | G→C (%) | 9 (69.2) | |
| Chrx:154535443 G→C | N (%) | 12 (92.3) | |
| Wild Type (%) | 11 (84.6) | ||
| Mutant (%) | 1 (7.7) | ||
| 485+37 (G→T) | N (%) | 10 (76.9) | |
| Wild Type (%) | 10 (76.9) | ||
| Mutant (%) | 0 | ||
| G1116A | N (%) | 12 (92.3) | |
| Wild Type (%) | 9 (69.2) | ||
| Mutant | G→A (%) | 2 (15.4) | |
| G→T (%) | 1 (7.7) | ||
| C563T | N (%) | 11 (84.6) | |
| Wild Type (%) | 11 (84.6) | ||
| Mutant | C→T (%) | 0 |
Note: four patients who had double mutation at their G6PD gene; (1st, G267+119C/T (GàC), and G1116A (GàT), 2nd, G267+119C/T (GàC), Chrx:154535443 (GàC), 3rd, A376G (AàT), G267+119C/T (GàC), and 4th, A376G (AàG), G267+119C/T (GàC)).
Table 3: Frequency of G6PD genetic mutations at Exon three to eleven among malaria suspected patients.
The average mean of phenotypic G6PD biosensor result of patients who had mutations was 1.79 (0.1-5.0) while the average mean of patients who hadn’t mutations was 0.87 (0.1-1.4) (mean diff. =0.9, 95% CI; -0.954-2.801, P=0.3) which was not statistically significant.
Among the sequenced samples for G6PD, five were positive for malaria parasites (four P. falciparum and one P. vivax). The binary logistic regression revealed no significant association between G6PD mutation and malaria-positive cases even if the odds of having mutation increase by 1.3 times in positive cases than in malaria-negative cases (COR, 1.3; 95% CI, 0.09-20.1; P=0.8).
Primaquine is the only available and effective drug of choice for the treatment of the pre-erythrocytic stage (liver stage) of P. vivax against relapses and the gametocyte stage of P. falciparum which is a key point for the transmission interruption of malaria parasites. However, it can cause acute hemolytic anemia in individuals with G6PD deficiency. Therefore, understanding the genetic influence of malaria susceptibility in humans is crucial for the elimination program [18].
This study revealed that the prevalence rate of G6PD deficiency was 4.8% using a quantitative rapid point-of-care CareStart biosensor machine which is moderate according to WHO reports [28]. This is comparable with, a study performed in Southwest Ethiopia around Gambela (average altitude of 300-500 meters above sea level) by Lo et al., and Tsegaye et al., revealed 4.3% and 7.3% respectively [1,12]. However, there are differences in the altitude and malaria endemicity pattern between both areas. Similarly, a review paper performed in Southwest Ethiopia stated that the G6PD deficiency prevalence was above 1-3% [17]. Relatively, broader parts of the country were included by Shitaye and his colleagues and revealed an overall prevalence rate of 1.4% G6PD deficiency [26]. Study conducted in Sudan indicated , the prevalence rate of G6PD deficiency was 13.1% using Point-of care-testing qualitative CareStart rapid diagnostic tests [29].
This study revealed that almost all G6PD deficient participants were above fourteen years old. This was the same as the study performed by Nguetse and his colleagues (P=0.29). Likewise, a study performed in Saudi Arabia also revealed that G6PD deficiency increased as age increased [12,30].
This study also revealed that males and females had an equally likely chance of being G6PD deficient (AOR, 0.8, 95% CI, 0.423-1.498, P=0.5). This was concordant with a study performed in Southwest Ethiopia (AOR, 1.4, P>0.05), in Sudan and northern Thailand [13,31]. Contrarily, a study conducted in Saudi Arabia and Egypt showed that males had a higher chance of being G6PD deficient than females. This might be true that since it is an X-linked disorder, males can have the chance of receiving the deficient gene during the inheritance of a single X-chromosome copy [13,30,32].
All malaria-positive cases had an insignificant association with G6PD deficiency despite the high odds value (AOR, 2.6; 95% CI, 0.47-14.26; P=0.3). Similarly, research performed by Getasew Shitaye and his colleagues revealed a negative association of G6PD deficiency with malaria parasites (P=0.9) [26]. By far, the natural selection pressure exerted by malaria parasites led to rising G6PD deficiency [12,33]. This disagreement could be due to the methodological difference.
The logistics regression output between the G6PD enzyme level and the parasite load was P=0.2, and the association was statistically insignificant. This is the same with Lo and her colleagues and contrarily with Tsegaye and his colleagues (P<0.0001) [1,13]. G6PD deficiency is associated with hypo-parasitemia [10,34-36]. Male hemizygous and female homozygotes had a high protective effect against severe malaria (P=0.0006), and cerebral malaria (P=0.0005) [37]. Male hemizygous and female heterozygotes have a reduced risk of cerebral malaria [38].
Of the participants, the common mutation, 376 (A → G and A → T) was detected in 3 (23.1%) participants. G267+119C/T (G → C) was detected in 9 (69.2%) while G1116A (G → A and G → T) was detected in 3 (23.1%) of the participants. In Southwest Ethiopia, a nationwide study revealed 6.1% and 8.9% of A376G genomic mutations respectively while G267+119C/T and G1116A gene mutations accounted for about 1.2% [1,18]. In this study, almost all participants had G267+119C/T (9) gene mutations than other variants. Of these, three of the participants were heterozygous whereas homozygous and hemizygous were equally three in each participant. One mutation in G1116T was also heterozygous while surprisingly, one heterozygous participant had a double mutation in her G6PD gene.
This study also revealed new nucleotide substitution at the position of 376 (A → T) and 1116 (G → T) in Ethiopia. Similarly, 376 (A → T) was first isolated in Mexico (126 Asn → Tyr) named San Luis Potosi [38]. This finding didn’t show G6PD A- (G202A) and Mediterranean (C543T) variants among sequenced samples. However, G6PD A- was a common variant in Africa [38] and in Ethiopia, 3.5% of G6PD A- was detected around Southwest Ethiopia [1] while no mutation was detected for the Mediterranean variant in Ethiopia.
This finding revealed a moderate prevalence rate of G6PD deficiency rate. In a place where the prevalence rate of male G6PD deficiency is greater than 3-5%, mass screening should be performed before administration. In the case of this finding, the clinicians have to consider the risk of hemolysis during primaquine treatment in the study area. Genotypically, almost all included study participants had G267+119C/T G6PD gene mutation, and rarely A376G and G1116A had been shown. Genotypic analysis with broader samples could be essential to identify the distribution of dominant variants in the study site. Since genetic mutation is a gradual and recurring situation, continuous assessment of a given population could be mandatory.
Ethics approval and consent to participate
Ethical approval was sought from the IRB of Aklilu Lemma Institute of Pathobiology, Addis Ababa University, and Ethiopian Public Health Institute after presenting the study proposal. Written informed consent was obtained from participants ages greater than eighteen years old and oral assent from all participants aged less than eighteen years old. All collected information was kept confidential and records were coded. No personal identifiers were kept in the database or used to report findings. The data was restricted using password-locked and personal computers other than the principal investigator while the hard copies were locked in the cardboard.
All authors read and approved the submission for publication.
The data produced in the study is included in the main manuscript and the rest are available upon reasonable request from the corresponding author.
The authors declare that they have no conflicts of interest.
The funding agents for this paper were the Global Fund through the Ministry of Health-Ethiopia, Addis Ababa University, Aklilu Lemma Institute of Pathobiology, Access Bio Inc. Company, and Ethiopian Public Health Institute.
MTN, SDu, LG, and SMF conceived the research idea and participated in the design of the study; MTN, EK, SDe, TTS, DN, BG, BM, and MY participated in the collection of samples; EL, DK, LW, MTN, TTS, and AAd participated in laboratory diagnosis and analysis of the data; MTN, SDe, LG, SMF, TTS, and AAd wrote the paper; LG, SDu, BD, AAb, SMF, HA, EL, DN, and GT participated in the data interpretation and revision of the manuscript.
First, we would like to thank Almighty God for giving me the strength to face and succeed in my life challenges. Secondly, we would like to provide my heart full thanks to my brother, Mr. Afework who is polite and cooperative and he converted the consent form, information sheet, and questionnaire into the Gammogna language version. Moreover, we would like to thank Malaria and the Neglected Tropical Disease Research Team for their unlimited support in the thesis write-up.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Negash MT, Golassa L, Dugassa S, Feleke SM, Nega D, Abebe A, et al. (2022) Genetic Variants among Malaria-Suspected Patients with Glucose-6 Phosphate Dehydrogenase Deficiency. Adv Tech Biol Med. 10:390.
Received: 01-Nov-2022, Manuscript No. ATBM-22-21012; Editor assigned: 04-Nov-2022, Pre QC No. ATBM-22-21012 (PQ); Reviewed: 11-Nov-2022, QC No. ATBM-22-21012; Revised: 25-Nov-2022, Manuscript No. ATBM-22-21012 (R); Published: 02-Dec-2022 , DOI: 10.35248/2379-1764.22.10.390
Copyright: © 2022 Negash MT, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.