Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2020)Volume 13, Issue 3
Genus Streptomyces has been a source of various clinically significant bioactive metabolites. Taxonomically, Streptomyces formicae KY5 is a new and different species. The complete genome sequences of S. formicae KY5 is available in the public DNA sequence databases for different analysis. The accessibility of the genomic sequence presents an excellent opportunity to explore the secondary metabolites potential of this distinct Streptomyces species. In this study, we employed the advance bioinformatics resources to annotate the total genome sequence of S. formicae KY5. Bioinformatics tools are applied to locate all the secondary metabolites hiding beneath their biosynthetic gene clusters (BGCs). The S. formicae KY5 is found to synthesis distinct and various secondary metabolites by undergoing the designated genomic encoding. Predictive analysis conveys that this strain has 34 gene clusters to encode potential secondary metabolites. For structural similarity with other drugs, we scanned the drug bank database, drug target and drug with the highest similarity was retrieved from PDB for molecular docking. Molecular docking analysis was carried out through molecular operating tool to evaluate drug-like potential of the chemical compounds. Three drugs like compounds were predicted from S.
Streptomyces formicae; Secondary metabolites; Gene clusters; In-silico
In the survival of microbial culture, natural products produced by these microbes play a significant role. These organic compounds aids the microbes in their interactions with the environment and with other organisms [1]. These natural products obtained from microorganisms play their impacting part in almost every biomedical field of life such as agriculture, healthcare, veterinary and nutritional fields [2]. Microbial natural products or secondary metabolites can be used as a pharmaceutical, nutritional and agricultural agents. These microbial natural products comprise of saccharides, terpenoids, non-ribosomal peptides (NRPs), polyketides (PKs), peptides which are post-transnationally modified (RiPPs), some hybrid natural products, and ribosomally synthesized compounds. These natural products are synthesized by biosynthetic gene clusters (BGC), only found in bacteria, fungi and plants. Biosynthetic gene cluster contain genes in the form of groups located physically at a single locus on microbial plasmid and chromosomal DNA. Secondary metabolites are synthesized by these groups of genes. These BGCs because of evolutionary basis hold rich genetic variety which provide a route to diversity in chemicals and their respective secondary metabolite [3,4]. To identify an unspecified natural metabolite we imply genome mining. With the help of genome mining, the gene clusters required for the production of natural metabolites in the genomic sequence of an organism can be investigated. A genome contains all the information necessary for an organism's production and growth in the form of genes that are accumulated in a set of DNA. From the entire database stored we can easily extract our interested BGC with the help of genome mining [5]. Streptomycetes, a gram-positive saprophyte is famous for their inherent talent to yield pharmaceutically relevant secondary metabolites. They produce more than half of all known antibiotics [6] and possess the potential to produce a considerable of 105 secondary metabolites according to an arithmetical study [7]. They are well known for producing various clinically important bioactive metabolites which include the antibiotics tetracycline and streptomycin, the anthelmintic avermectin, the antifungal amphotericin B, the antitumor mitomycin C, the immunosuppressant rapamycin and tacrolimus [8]. S. formicae KY5 (Bacteria; Actinobacteria; Actinobacteria; Streptomycetales; Streptomycetaceae; Streptomyces; Streptomyces formicae) is recently discovered species, extracted from an African plant named ant Tetraponera penzigi [9,10]. S. formicae KY5 is a talented species because of its ability to produce new natural products. S. formicae KY5 is revealed to produce an unspecified compound which is antifungal and active against human pathogen Lomentospora prolificans, a multi-drug resistance, and formicamycins, a pentacyclic polyketides group, which show effective antibacterial actions against vancomycin-resistant Enterococci (VRE) isolates and clinical methicillin-resistant Staphylococcus aureus (MRSA), but this species does not show activity against gram-negative microorganism. It is estimated that at least 45 additional natural products are encoded by S. formicea KY5 [11]. Due to misuse of antibiotics bacteria and infection causing agents becoming actively resistant to their respective drugs [12]. We have selected S. formicea species because of its potential to encode various secondary metabolites. In this study, we used various computational tools to mine publically accessible entire genome sequence of S. formicae KY5 to investigate the gene coding potential of this species for secondary metabolites. The analytical scaffold (initial structure leading to the drug) of putative natural products of S. formicae KY5 recognized are further investigated by employing chemoinformatics resources to perceive the potentials of compounds similar to drugs.
Retrieval of genomic sequence
The complete genome sequence of S. formicae KY5 was retrieved from NCBI Gene bank (accession number: NZ-CPO22685) on May 9, 2018.
Natural products and secondary metabolites prediction We have started with the analysis of its genome through a web server namely AntiSMASH [13], specially designed for investigation of the secondary metabolites coding BCGs. The updated version of the AntiSMASH (4.0) [14] uses a constructive database of currently known BGCs from the MIBiG (Minimum Information about a Biosynthetic Gene cluster) to pinpoint the all the secondary metabolite gene coding clusters. AntiSMASH uses CASSIS and SANDPUMA algorithm to predict the boundaries of gene clusters and to enhanced substrate specificity predictions of secondary metabolites respectively. The most recent AntiSMASH version is incorporated with Cluster Finder and Cluster Blast modules to the unidentified types of BGCs by resembling them with similar identified BGCs.
Elucidation of chemical structure
The chemical structures of the secondary metabolites as predicted through antiSMASH were elucidate using ChemDraw chemoinformatics tool [15]. The chemical structures were converted to SMILES format for further analysis.
Screening through drug bank database
The predicted secondary metabolites were analyzed to find out their respective drug targets and homolog drug-like chemical scaffolds identification using Drug bank resource [16].
Drug-like potential evaluation by molecular docking
The molecular operating tool or MOE (v.2016) tool was used to predict the putative secondary metabolites capability to act like a drug on the account of the vital Lipinski rule of five [17]. To understand the orientation and binding of secondary metabolites with respective drug target, we look upon the MOE tool to calculate the molecular docking Scores, binding energies, and binding affinities.
Natural products harboring potential of S. formicae KY5 To evaluate the potential of secondary metabolites pathways in S. formicae, we have employed antiSMASH version 4 that predicted at least 34 biosynthetic gene clusters. Among many prediction models implemented, the antiSMASH is a reliable, inclusive, powerful and a computational course for the estimation and explanation of recognized and unidentified biosynthetic gene clusters using probabilistic algorithm [18]. Genome data scanning of S. formicae reveals different secondary metabolites encoded by BGCs. A single type 2 PKS secondary metabolite that codes for formicamycins (10), three types 1 PKS, eleven BGCS codes for NRPS and six for terpene, in which one terpene is similar to antibiotic albaflavenone produced by Streptomyces coelicolor [19] and two BGCs are found to encode siderophore in which one show 100% similarity with desferrioxamine.
Secondary metabolites encoded by some of the BGCs in S. formicae KY5
Non-ribosomal peptides: The analysis of S. formicae KY5 genome sequence disclose their capability to produce multiple sets of NRPs. Nonribosomal peptides (NRPs) are used as drugs, toxins, pigmentations agents and siderophores. A multi-domain enzyme which is a nonribosomal peptide synthetase (NRPs) complex manifest NRPs [20]. AntiSMASH resource identifies 11 BGCs codings for NRPs from the analysis of the genome used here.
Polyketides: S. formicae KY5 is found to have a gene cluster for a single type 2 polyketide synthetase which is involved in the biosynthesis of the formicamycins [9]. Biosynthesis of bioactive polyketides natural compounds which are also known as Polyketides Synthetases (PKS) is ordered by a multi-domain enzyme complex. During genome analysis of S. formicae KY5 antiSMASH resources identified BGCs encoding three types of PKS. In medicines, polyketides metabolites play a significant role and been commonly used to cure delicate and degenerative diseases. The type 1 polyketide synthetase is arranged into components that catalyze repetitively on the reduced polyketides elongations and chain synthesis, for example, erythromycin. Only one set of repetitively acting cyclic polyketides biosynthetic domains is fetched by type II Polyketide synthetase, such as tetracenomycin C. ACPs (acyl carrier proteins) are employed by type I and II PKS to make acyl Co A substrate active for the development of polyketides intermediates.
Bacteriocins: Bacteria and archaea species yield some ribosomally synthesized peptides of lower molecular weight which shows antimicrobial activities and are termed as bacteriocins. AntiSMASH identified one bacteriocin BGCs during the genome analysis of S. formicae KY5. Most of the food preservatives and antibiotics being used are bacteriocins [21,22] and there are 4 sub-groups of bacteriocins due to diversity in the method of action and chemical structure. Bacteriocins of class 1 show antimicrobial activities and action like post transitionally modified peptides such as sactipeptides, lantipeptides and lasso peptides [23].
Terpenes: AntiSMASH analysis of S. formicae KY5 identified six BGCs which encodes for terpenes. One of the encoded terpene show 100% similarity with the antibiotic made by Streptomyces coelicolor, albaflavenone [19]. Fungal and plant metabolites are commonly known as terpenes and a few odoriferous terpenes are predicted so far from certain bacteria. Terpenes made from bacteria are reported to be antimicrobial in nature and used to aid in the manufacture of precise substances such as fermentations [24]. During genome analysis of formicae KY5 a single cluster for ectoine type of secondary metabolites predicted which shows 100% similarity with the biosynthetic gene cluster of ectoine. The natural cell-protecting compatible solute or ectoine behaves as an osmolyte that enables the survival of bacterium species in concentrated salty ecological stress [25].
Siderophores: During genome analysis of S. formicae KY5 there are at least two siderophores BGCs, in which one siderophore BGC show 83% similarity with desferrioxamine B. Plants growing in an iron-deficient environment and certain microorganisms produce organic compounds with low molecular weights, these organic compounds are known as siderophore. Siderophore is diverse in chemical structure and other properties but the most significant applications are that they hold the talent to hold together a variety of metals in additions to iron [26]. Streptomyces pilosus produce a siderophore named desferrioxamine B, which is marketed as the mesylate salt with the trade name “Desferal” and its action is to remove the excess of iron resulting from the supportive treatment of thalassemia. It is essential to inject the drug, but, an oral replacement is also required [27]. A single cluster for melanin type of secondary metabolite is predicted through antiSMASH during genome analysis of S. formicae KY5. The dark pigments are synthesized and excreted by most of the streptomyces species. Various microorganisms produce melanin by fermentative oxidations. Radioprotective antioxidant properties of melanin are reported to protect living organisms from ultraviolet radiation. Therefore, melanin is specially used in cosmetic products and in pharmacology and medicines [28]. A single cluster for each thiopeptide and butyrolactone like products are predicted to encode by Streptomyces formicae KY5 through antiSMASH during genome analysis. Highly enhanced, sulfur-containing macrocyclic peptides make a family of thiopeptide. A significant class of natural products is represented by thiopeptides resulting in ribosomally synthesized and post-translationally modified peptides. One of the typical thiopeptide antibiotics is Cyclothiazomycin [29]. Various classes of diffusible signaling molecules that activate secondary metabolites biosynthesis are produced by certain Streptomyces bacteria such as the biosynthesis of gamma-butyrolactone [30]. Butyrolactone is category of compounds that embrace A-factor and other signaling agents from Streptomyces spp [31].
Hybrid BGCs: S. formicae KY5 genome analysis through antiSMASH revealed a number of hybrid BGCs including two T3pks-Nrps, Ladderane-T1pks-Nrps, T3pks-lantipeptidelessopeptide, bacteriocin-T3pks-lantipeptide-Nrps, blactam-T1pks, lantipeptide-T1pks-Nrps, Nrps-Ladderane, Amglyccycl-terpene- Nrps, oligosaccharide-T1pks-nrps, and lantipeptide-T1pks. Amglyccycl-terpene-Nrps show 55% similarity with a cyclic depsipeptide antibiotic, Telomycin show antibacterial activity against gram positive bacteria [32]. Lantipeptide-T1pks show 46% similarity with Abyssomicin BGC which is reported to be active against methicillin-resistant Staphylococcus aureus (MRSA) and other gram positive bacteria [33]. Lantipeptide-T1pks also possess drug-like potential, it follows Lipinski rules of five and drug bank database scanning revealed that it holds structural similarity with (8R, 9Z, 12Z)-8-hydroxy-6-oxooctedeca-9, 12-dienoic acid.
Drug like potentials of putative secondary metabolites
Aside from metabolites that are high in molecular weight, three compounds (Figure 1) were predicted as a potential drug like from the genomic sequence of S. formicae KY5. Compound 1 and 2 (Table 1) encodes from Nonribosomal peptides while compound 3 (Table 1) is encoded by hybrid BGC lantipeptide-type 1 PKs. The potential of these compounds to act as drug like is demonstrated by the essential Lipinski rules of five [17]. These metabolites have the molecular weight of not more than 500 Daltons and the number of acceptors in hydrogen bond is less than 10 while and hydrogen bond donor are less than 5. Similarly the value of log p (octanol-water partition coefficient) is not greater than 5. Detail of these properties of compound A, B and C are given in Table 1. For further investigations of the drug-like the potential of these secondary metabolites, firstly the drug bank database was scanned [34]. Drug Bank database scanning reveal that the Compound-A hold structural similarity with (8ar)-Hexahydropyrrolo [1, 2-a] Pyrazine-1, 4-Dione which lies in an experimental group of drugs according to the drug bank database. The target of this compound is a protein called Chitinase B, present in a pathogenic bacteria Serratia marcescens. S. marcescens is concerned with a lot of serious infections such as pneumonia [35], infections of lower respiratory tracts [36], infections of urinary tract [37], wound infections, bloodstream infections and meningitis [38]. S. marcescens is also an important cause of ocular infections with high prevalence in contact lens-related keratitis [39,40]. The compound 1 reported in this study could be a potential candidate for future studies against infections caused by S. marcescens.
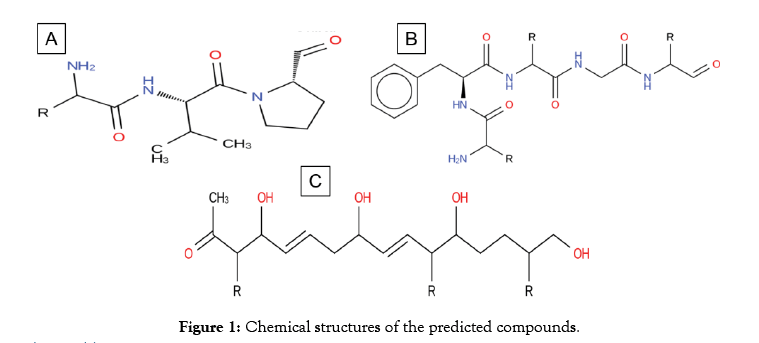
Figure 1: Chemical structures of the predicted compounds.
| Compound A | Compound B | Compound C | |
|---|---|---|---|
| Property | Value | Value | Value |
| Weight | 269.34 | 419.48 | 342.48 |
| Log P | 0.24 | 0.30 | 1.93 |
| Log S | -1.70 | -2.84 | -2.32 |
| Hydrogen Donor | 2 | 5 | 4 |
| Hydrogen acceptor | 4 | 6 | 5 |
| Toxic | No | No | No |
Table 1: The potential properties of compound A, B, and C to act like a drug.
Compound-B shows structural similarity with oxetacaine. Oxetacaine, also called oxethazaince is potent local anesthetic [41]. Its activity has revealed to conceal dysphagia, relieve pain caused by chronic gastritis, duodenal ulcer and reflux [42]. Oxetacaine is found to have a local anesthetic effect on the gastric mucosa [41]. It is a part of antacids that escalate the pH of gastric while accommodating relief from pain at a lower dosage for a longer duration. This ability of oxetacaine is reported to conceal the hyperacidity symptoms [43]. Oxetacaine is describing to generate a reversible loss of sensation and to accommodate a prompt and continued relief of pain. The testified derivatives of the EMA which produce herbal contained oxetacaine.
Predicted Compound-C have structural similarity with (8R, 9Z, 12Z)-8-hydroxy-6-oxo octadeca-9, 12-dienoic acid or linoleic acid. It is one of the essential fatty acids must be obtained for normal growth and for health. Linoleic acid is involved in the synthesis of arachidonic acid, hence engaged in the production of signaling molecules such as prostaglandins, thromboxane and leukotriene. The deficiency of linoleic acid can lead to many disturbances in standard growth and matureness of an organism [44] (Figure 2). To additionally understand the receptor binding potential of putative Compound-A-C (Table 2), we apply the molecular docking analysis. For putative compound 1, Chitinase B, the complex structure is derived from protein data bank (PDB) (ID: P11797) with the motive of docking. Similarly, for putative compounds 2 and 3 (Table 2), the protein structures of gastrin (ID: P01350) and Peroxisome proliferator-activated receptor gamma (ID: P37231) are downloaded respectively. The best-docked poses for each of the three Compounds (Table 2) show favorable interaction with important residues. Best docking scores, binding energies and binding affinities are exposed by putative Compound-1, 2 and 3 (Table 2 and Figure 2) after docking with their respective targets. S. formicae KY5 contains a diverse set of natural products (secondary metabolites). One of the main challenges of modern biotechnology is the discovery of novel medicines because of the emergence of drug resistance among pathogenic bacteria. It is need of the time to explore novel alternate antimicrobial natural compounds significant for modern medicines. These coding clusters of the prospective secondary metabolites propose valuable targets for experimental examination to develop new resources.
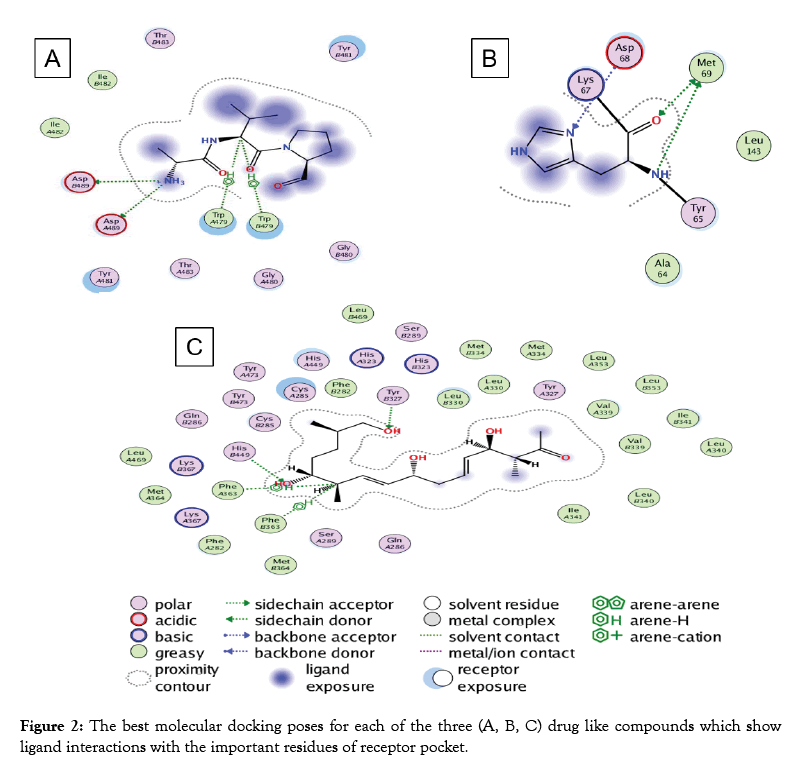
Figure 2: The best molecular docking poses for each of the three (A, B, C) drug like compounds which show ligand interactions with the important residues of receptor pocket.
| Ligand | Receptor | Docking score (S) | RMSD-refine | E-conf | E-place | E-score | E-refine |
|---|---|---|---|---|---|---|---|
| Compound A | 1HOG | -8.35 | 3.1 | 20.6 | -41.5 | -6.9 | -21.6 |
| Compound B | 5WRJ | -12.95 | 1.5 | -19.1 | -82.3 | -12.3 | -42.5 |
| Compound C | 2ATH | -12.50 | 1.6 | 10.6 | -80.4 | -10 | -28 |
Table 2: The best molecular docking scores for each drug like compound after docking with their respective drug targets.
We have analyzed the genome of the S. formicae KY5 which resulted in the prediction of 34 BGCs. These 34 BGCs gave us 16 compounds out of which 3 compounds follow Lipinski rule of five and show similarity with certain drugs in the drug bank database. We docked our putative compounds with the drug targets of the highest similar drug through a molecular operating tool that show the best docking score. An additional practical approach is required to convert these biosynthetic gene clusters into corresponding products to test their drug-like behavior for future new drug discovery.
The authors declare that there is no conflict of interest.
Authors are thankful to Bahauddin Zakariya University, Multan, Pakistan for providing the necessary infrastructure for conducting this study.
Citation: Shah M, Gul S, Amjad A, Javed MS, Fatima B, Nawaz H, et al. (2019). Genome Mining of Streptomyces formicae KY5 for Potential Drug like Natural Products Characterizations. J Proteomics Bioinform 12:7. doi: 10.35248/0974-276X.19.12.505
Received: 14-Oct-2019 Accepted: 02-Dec-2019 Published: 09-Dec-2019 , DOI: 10.35248/0974-276X.19.12.505
Copyright: © 2019 Shah M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.