Research Article - (2023)Volume 9, Issue 1
Greatest Common Divisor Deciphered SARS-CoV-2 Superpandemic
Ke Jia Dong1* and Wen Hua Li2Abstract
Global cases and deaths showed that COVID-19 and 1918 influenza were not normal pandemics, but superpandemics. Was there a common mechanism to push pandemics into superpandemics? This common ground was explored here from a new perspective and approach using the Greatest Common Divisor (GCD). The results showed that superpandemic viruses played tricks like superbugs. One subtlety was that SARS-CoV-2 fighted antibodies, just as superbugs fighted antibiotics. The SARS-CoV-2 spike and ORF8 proteins recognized the “Achilles heel” of secretory antibodies, namely J-chains and secretory components, and hijacked them respectively. Another subtlety was that SARS-CoV-2 expanded ORF8 protein as a superpandemic catalyst, just as drug-resistant enzyme facilitated the spread of superbugs. The SARS-CoV-2 ORF8 protein corresponded to the 1918 H1N1 virus neuraminidase. Both functioned as glycosylated-modification enzyme and RNA base-modification enzyme. Tampering with the enzymes was not found in SARS-CoV and pandemic (H1N1) 2009 virus. The synergy of spike and ORF8 proteins acted as the ignition for SARS-CoV-2 superpandemic. Through GCD analysis of clinical and experimental results of different coronaviruses, it was proposed to comprehend the epidemiological traceability and evolutionary from virus sequence up to virus GCD. We sincerely recommend the GCD platform to WHO for early warning.
Keywords
COVID-19; SARS-CoV-2; Virus; Greatest common divisor
Introduction
The number is life. So far, more than 6.6 million deaths of COVID-19 have been confirmed worldwide. The number of deaths was similar to that of the 1918 flu [1]. While the two genomes were markedly different, SARS-CoV-2 was as devastating a pandemic as the 1918 flu virus [1,2]. The prevalence indicated that neither COVID-19 nor the 1918 flu were normal pandemics, but superpandemics. It was known that the transformation of antigen from drift to shift turned virus epidemic into pandemic, so what was the molecular basis for the virus to go further from the pandemic to a superpandemic? At present, viral metagenomics, protein structure and genetic manipulation were the three fundamental approaches for our understanding of viral biology [3]. However, the key issue was the complex relationship between protein structure and function. Although more than 200 million protein structure predictions were available in the AlphaFold database, structure similarity did not necessarily equal function similarity. In particular, the structural and functional information of intrinsically disordered protein extended the traditional structure-function pattern of protein. Some new strategies were needed to decode SARS-COV-2. In the vast universe, number law showed up in everything, such as from the redshift of cosmic microwave background radiation to the number of C. elegans cells, from the quantum number of particle physics to the life cycle of periodic cicadas [4,5]. These studies revealed that number theory could bridge the three fields of mathematics, physics and biology [6,7]. Since the process of finding the Greatest Common Divisor (GCD) was the process of finding common ground, the protein@integer GCD platform was created here to uncover the common mysteries of superpandemic viruses. Our research showed that superpandemic viruses differed from pandemic viruses in that they expanded a self-modification multifunctional enzyme, which acted as a catalyst for virus superpandemic, just as enzyme contributed to the spread of superbugs. Through combined computational analysis with clinical epidemiological experimental research, the superpandemic mechanism is proposed, and the greatest common divisor is raised from the original mathematical term to the epidemiological concept. The GCD platform was not only used to crack the superpandemic mechanism, but also used to capture superpandemic strains of different coronaviruses.
Materials and Methods
Integer theory was proposed to study the structure and function of proteins. Based on the standard amino acids mass and the common characteristics, protein was encoded into a vigesimal system. The cardinal number of the vigesimal system corresponds to the number of standard amino acids. The protein sequences were encoded into digital sequences, and performed ab initio calculations using the vigesimal system. Protein integers were large integers. Then, the protein functions were decoded by integer theory, such as large integer factorization or Greatest Common Divisor (GCD). The GCD was originally a common mathematical term, and referred to the largest of the common divisors of two or more integers. Here, extending to the field of molecular biology, protein GCD referred to the common function between two or more different proteins, which reflected the corresponding function or functional synergy between different proteins. It could be understood from two perspectives: one was to find the GCD from the perspective of a single proteome, that is, to find corresponding function or functional synergy of different proteins from the proteome; The other was to find the GCD from the perspective of multiple proteomes, that is, to find the domination or driving function of different interactome from multiple proteomes. Here based on the theorem of GCD, a protein GCD platform was constructed and further applied it to the pathogenesis analysis of different pathogens. Protein GCD platform was divided into three steps: the first step was to encode the protein as an integer, namely protein@integer; the second step was to calculate the protein GCD from protein@integer databank; the third step is to analyze pathogenic mechanisms from protein GCD database. The GCD value greater than radix 20 was a significant criterion for protein GCD analysis. Understanding the global properties and functions of proteins through integer factorization and GCD provided a bridge between molecular biology and number theory (Figure 1).
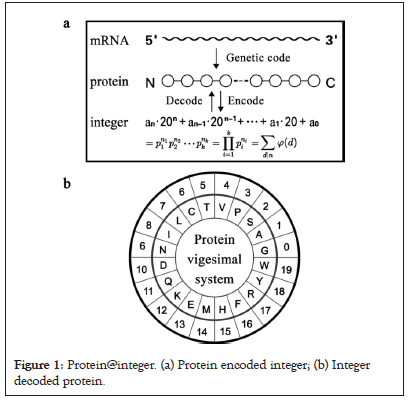
Figure 1: protein@integer. (a) Protein encoded integer; (b) Integer decoded protein.
Data availability
Virus proteome were obtained from NCBI and GISAID database. All data supporting the findings of this study are available in the text and supplementary information.
Note: SARS-CoV-2 (NC_045512.2); SARS-CoV (NC_004718.3); A/Brevig Mission/1/1918(H1N1); A/California/07/2009(H1N1); S. aureus (RBAU01000015.1 and CP010944.1); S. pneumoniae (AE007317.1); H. influenzae (AP022867.1)
Code availability
1. protein@integer software
2. protein@integer test and demo
Results and Discussion
The GCD decoded virus superpandemic
We sought to determine whether different superpandemic pathogens shared an identical or similar mechanism. In order to find the common mechanism, we developed the GCD platform to search for the molecular basis from different pandemic viruses and superbugs (Figure 1a). The key to the molecular basis was to identify the common function of different antigens. Through GCD analysis of the SARS-CoV-2 proteome versus pandemic influenza virus and superbug antigens, we identified the S and ORF8 proteins as key antigens causing the SARS-CoV-2 superpandemic (Supplementary Tables 1 and 2). The GCD results showed that the key antigens of SARS-CoV-2 had the dual properties of immune and enzyme. On the one hand, the SARS-CoV-2 S protein shared the GCD with the clumping factor A (ClfA) of Methicillin-Resistant Staphylococcus aureus (MRSA) and hijacked the SIg J chain (SIg-JC); the SARS-CoV-2 ORF8 protein shared the GCD with the SpsA or PspA of Methicillin-Resistant Streptococcus pneumoniae (MRSP) and hijacked the SIg secretory component (SIg-SC) (Figure 1b and Supplementary Tables 1 and 3). On the other hand, the ORF8 protein of SARS-CoV-2 corresponded to the neuraminidase of the 1918 H1N1 virus (Supplementary Table 1). The ORF8 protein and the neuraminidase shared the GCD not only with the human glycosylation-modified enzymes GlcNAcase and HexNAcase, but also with the human RNA editing enzymes ADAR1 and ADAR2. Both might function as glycosylation-modified enzymes and RNA base-modified enzymes. In contrast, tampering with the enzymes was not found in SARS-CoV and the pandemic 2009 H1N1 virus (Supplementary Table 4). These GCD results explained why SARS-CoV-2 possessed the superpandemic characteristcs of the 1918 flu virus.
Next, we further investigated how SARS-CoV-2 exerted a synergistic effect of immunohijacking and enzyme modification. The key to synergy was to identify common molecular pathways. We used the GCD to detect whether there was a common molecular pathway between SIg/ SC hijacking and enzyme modification, and found that SARS-CoV-2 worked together to destroy CD4 cells through two different pathways, namely ORF8-SIg/SC-CD4 and ORF8-ADAR1-CD4. Since ADAR was also RNA base-modified enzyme, we used the GCD to analyze SARS-CoV-2 accessory proteins and nonstructural proteins. The GCD results showed that the accessory protein ORF8 shared the GCD with the nonstructural proteins nsp1, 11,13 (Supplementary Tables 5 and 6). This result indicated that the synergistic interaction of modification enzyme ORF8 and helicase nsp13. The SARS-CoV-2 ORF8-based synergistic effects told us that although superpandemic viruses still lacked protein translation system, they had evolved a unified mechanism of replication, immunity, and modification through the expansion of single-functional enzymes to multifunctional enzymes.
Finally, we used the unified mechanism to decode the emergence of SARS-CoV-2 variants of concern. Why was Omicron, the SARS CoV-2 variant, so subtle? In Supplementary Tables 7 and 8, Omicron, such as OQ151515, showed that it had special properties similar to the original strain but distinct from the other variants. Omicron S and ORF8 proteins had synergistic immune and modification effects.
The GCD capturing superpandemic virus
Table 1 took SARS-CoV-2 and the 1918 H1N1 virus as standard superpandemic viruses and compared them with human, bat and synthetic coronaviruses. The S and ORF8 proteins of SARS-CoV-2 or the hemagglutinin and neuraminidase of the 1918 H1N1 virus were used as reference antigens. In human coronaviruses, the GCD results showed that HCoV-OC43 and SARS-CoV-2 S protein had the GCD, indicating that HCoV-OC43 and SARS-CoV-2 S protein had similar functions. In bat coronaviruses, the BtCoV-HKU3-1 ORF8 protein and SARS-CoV-2 ORF8 protein had the GCD, indicating that the BtCoV-HKU3-1 ORF8 protein and SARS-CoV-2 ORF8 protein had similar functions. The S and ORF8 proteins of both RaTG13 (MN996532.2) and RpYN06 (MZ081381.1) did not have significant GCD with the SARS-CoV-2 S and ORF8 proteins or the 1918 H1N1 viral hemagglutinin and neuraminidase. However, the synthetic CoV (Bat-SRBD) S protein and ORF8 protein possessed the GCD with SARS-CoV-2 S protein and ORF8 protein, respectively. Therefore, although RaTG13 and RpYN06 are closer to SARS-CoV-2 than Bat-SRBD in sequence similarity, Bat-SRBD is more likely to become a superpandemic strain than RaTG13 and RpYN06 due to the function similarity (Table 1).
| GCD | SARS-CoV-2 | 1918 H1N1 Virus | |||
|---|---|---|---|---|---|
| S | ORF8 | H | N | ||
| Human CoV | NL63 S | 1 | 1 | 1 | 1 |
| 229E S | 1 | 1 | 1 | 1 | |
| OC43 S | 285 | 2 | 2 | 2 | |
| HKU1 S | 5 | 2 | 2 | 2 | |
| MERS S | 5 | 1 | 1 | 1 | |
| SARS S | 15 | 1 | 1 | 1 | |
| Bat CoV | RaTG13 S | 5 | 1 | 1 | 1 |
| ORF8 | 1 | 8 | 8 | 4 | |
| RpYN06 S | 5 | 1 | 1 | 7 | |
| ORF8 | 1 | 8 | 8 | 4 | |
| HKU3-1 S | 15 | 1 | 1 | 1 | |
| ORF8 | 1 | 1768 | 8 | 68 | |
| Synthetic CoV | Bat-SRBD S | 95 | 17 | 1 | 17 |
| ORF8 | 1 | 1768 | 8 | 68 | |
Note: 1. Bat-SRBD is more likely to be a superpandemic virus; 2. The recombinant bat HKU3-1 CoV (human OC43 CoV S protein instead of bat HKU3-1 CoV S protein) will become a new superpandemic virus.
Table 1: The GCD capturing super pandemic virus.
protein@integer it is a new model for studying proteins. By upgrading protein sequences to protein integers, we strove to decode virus superpandemic and capture superpandemic virus from a new perspective-GCD, unveiling the epidemic evolutionary mechanism (Figures 2 and 3).

Figure 2: Epidemic evolutionary mechanism.
Figure 3: Protein@Integer GCD. (a) Protein@Integer GCD platform. The GCD analyzed superpandemic virus and superbug bacteria; (b) SIg-JC/SC-hijacked pathway.
The GCD showed that superpandemic viruses and superbug bacteria found the Achilles heel of SIg-J Chain (JC) and Secretory Component (SC), which resulted in the SIg-JC/SC-hijacked immune invasion. The virus superpandemic possessed the superbug-like mechanism of multi-resistant Staphylococcus aureus and Streptococcus pneumoniae.
Firstly, the GCD results explained why SARS-CoV-2 possessed the superpandemic characteristics of the 1918 flu virus. The enigma was that SARS-CoV-2 ORF8 protein was not only the key antigen, but also a multifunctional modification enzyme, which corresponded to the neuraminidase of the 1918 H1N1 virus [8,9]. Both functioned as glycosylation-modified enzymes and RNA base-modified enzymes. Tampering with the enzymes was not found in SARS-CoV and the pandemic 2009 H1N1 virus. Second, for SARS-CoV-2, the key antigens S and ORF8 had the adhesin function like the multidrug-resistant S. aureus ClfA antigen and the S. pneumoniae PspA antigen, respectively [10-13]. These adhesin antigens hijacked SIg-JC or SC to enhance immune invasion. This mechanism could explain the T-cell pathology in COVID-19. Although T cells lacked ACE2 and AQP1 (Supplementary Table 2), T cells might be destroyed by the SARS-CoV-2 hijacked ORF8-SIg/SC-CD4 and ORF8-ADAR1-CD4 pathway [14], which was similar to that S. pneumoniae hijacked SIg SC to result in functional impairment of CD4+cells.
But how did the SARS-CoV-2 ORF8 protein became a multifunctional modification enzyme? The versatility of proteins was related to the self-assembly of proteins. The crystal structure showed that SARS-CoV-2 ORF8 protein had the same self-assembled dimer as GlcNAcase and HexNAcase [15-17]. These structures revealed why dimerization was crucial for catalytic activity. The dimeric form of ORF8 protein had been observed in the tobacco BY2 cell expression system [18]. ORF8 protein from single molecule to functional architecture showed the relationship between protein function evolution and self-assembly. The evolution of ORF8 protein self-assembly further expanded its versatility. Just as gene evolution from scratch had expanded protein diversity [19]. It will be a new antiviral strategy to regulate the functional evolution of viral protein by controlling it’s self-assembly. However, the enzymatic transition-state of ORF8 oligomer remained to be elucidated by further experiments.
The GCD results revealed a unified relationship for superpandemic virus replication, immunity and modification. Why was SARS-CoV-2 mutation and evasion so fast? Based on the GCD results, on the one hand, the intrinsic interaction between the accessory protein ORF8 and the nonstructural proteins nsp1, 11, 13 promoted SARS-CoV-2 replication. On the other hand, the accessory protein ORF8 and the nonstructural proteins nsp1, 11, and 13 participated in the dual-cycle of SARS-CoV-2 life cycle and glycan cycle by hijacking SIg-SC and modification enzyme. The SIg SC-hijacked and IgG Fc- glycan hydrolysis reduced antibody-mediated neutralization and opsonization, facilitating the synergistic effect of the accessory protein ORF8 and the nonstructural proteins nsp1, 11, and 13 to enhance virus replication, generate mutations and evasion [20]. The clinical observation of SARS-CoV-2 confirmed that the deletion of ORF8 gene broke this epidemiological evolution and returned the SARS-CoV-2 superpandemic to a normal pandemic, similar to the deletion of neuraminidase in influenza C virus.
In terms of prediction and early warning, the GCD platform enabled us from virus sequence up to virus GCD to comprehend epidemic principle. Although RaTG13 and RpYN06 are closer to SARS-CoV-2 than Bat-SRBD in sequence similarity, the GCD results support that Bat-SRBD is more likely to be a superpandemic virus than RaTG13 and RpYN06. Previous experiments had also shown that Bat-SRBD was infectious in cultured cells and in mice [21]. According to the GCD analysis in Table 1, among human CoVs, the results showed that human OC43 CoV S protein and SARS-CoV-2 S protein have significant GCD. Although human OC43 CoV and SARS-CoV-2 S proteins share 29% sequence similarity, the GCD effect reflects the common function of human OC43 CoV and SARS-CoV-2 S proteins [22]. Similarly, bat HKU3-1 CoV ORF8 and SARS-CoV-2 ORF8 proteins have significant GCD, reflecting the common function of bat HKU3-1 CoV and SARS-CoV-2 ORF8 proteins. These GCD results, on the one hand, were supported by existing experimental studies, and on the other hand, further indicated that recombinant bat HKU3-1 CoV (OC43 CoV S protein+HKU3-1 CoV) or recombinant human OC43 CoV (OC43 CoV+SARS-CoV-2 ORF8 protein) would become a new superpandemic virus.
Conclusion
In summary, it is necessary to establish the Superpandemic Elements Database (SPED) for in-depth research. We would like WHO to organize the WHO@GCD terrace to capture superpandemic strains from different kinds of pathogens, including synthetic and semi-synthetic ones. It is conducive to faster and earlier response to global emerging infectious diseases. May the suffering end and the world usher in light.
Acknowledgement
We thank the people fighting COVID in the world.
Author Contribution
Conceptualization, methodology, and computation: K.J.D; BigNumer software: W.H.L. and K.J.D.
Competing Interest
The authors declare no competing interests.
Additional Information
Supplementary Tables 1-8.
Supplementary information.
References
- World Health Organization.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270-273.
- Johnson BA, Graham RL, Menachery VD. Viral metagenomics, protein structure, and reverse genetics: Key strategies for investigating coronaviruses. Virol. 2018;517:30-37.
- Fisenko AI, Lemberg V. On the radiative and thermodynamic properties of the cosmic radiations using COBE FIRAS instrument data: I. Cosmic microwave background radiation. Astrophys Space Sci. 2014;352:221-230.
- Sulston JE. C. elegans: the cell lineage and beyond. Biosci Rep. 2003;23(2-3):49-66.
- Maddox J. Möbius and problems of inversion. Nature. 1990;344:377.
- Balasubramanian K, Gupta SP. Quantum molecular dynamics, topological, group theoretical and graph theoretical studies of protein-protein interactions. Current topics in medicinal chemistry.2019;19(6):426-443.
- Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 2008;82(21):10493-501.
- Nunthaboot N, Rungrotmongkol T, Malaisree M, Kaiyawet N, Decha P, Sompornpisut P, ET AL. Evolution of human receptor binding affinity of H1N1 hemagglutinins from 1918 to 2009 pandemic influenza A virus. J Chem Inf Model. 2010;50(8):1410-1417.
- Herman-Bausier P, Labate C, Towell AM, Derclaye S, Geoghegan JA, Dufrêne YF. Staphylococcus aureus clumping factor A is a force-sensitive molecular switch that activates bacterial adhesion. Proc Natl Acad Sci. 2018;115(21):5564-5569.
- Palmqvist N, Patti JM, Tarkowski A, Josefsson E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 2004;6(2):188-195.
- Sixbey JW, Yao QY. Immunoglobulin A-induced shift of Epstein-Barr virus tissue tropism. Sci. 1992;255(5051):1578-1580.
- Hammerschmidt S, Talay SR, Brandtzaeg P, Chhatwal GS. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol. 1997;25(6):1113-1124.
- Xufeng R, Nie D, Yang Q, Wang W, Cheng T, Wang Q. RNA editing enzyme ADAR1 is required for early T cell development. Blood sci. 2019;1(03):196-201.
- Mayer C, Vocadlo DJ, Mah M, Rupitz K, Stoll D, Warren RA, et al. Characterization of a β‐N‐acetylhexosaminidase and a β‐N‐acetylglucosaminidase/β‐glucosidase from Cellulomonas fimi. FEBS Lett. 2006;273(13):2929-2941.
- Mark BL, Mahuran DJ, Cherney MM, Zhao D, Knapp S, James MN. Crystal structure of human β-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay–Sachs disease. J Mol Biol. 2003;327(5):1093-109.
- Elsen NL, Patel SB, Ford RE, Hall DL, Hess F, Kandula H, et al. Insights into activity and inhibition from the crystal structure of human O-GlcNAcase. Nat Chem Biol 2017;13(6):613-615.
- Imamura T, Isozumi N, Higashimura Y, Ohki S, Mori M. Production of ORF8 protein from SARS-CoV-2 using an inducible virus-mediated expression system in suspension-cultured tobacco BY-2 cells. Plant Cell Reports. 2021;40:433-436.
- Zhang L, Ren Y, Yang T, Li G, Chen J, Gschwend AR, et al. Rapid evolution of protein diversity by de novo origination in Oryza. Nat Ecol Evol. 2019;3(4):679-90.
- Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci. 2021;118(2):e2021785118.
- Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci. 2008;105(50):19944-19949.
- Guo L, Wang Y, Kang L, Hu Y, Wang L, Zhong J, et al. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: A retrospective study. Emerg microbes & infect. 2021;10(1):664-76.
Author Info
Ke Jia Dong1* and Wen Hua Li22School of Computer Science and Technology, Hainan University, Hainan, China
Citation: Dong KJ, Li WH (2023) Greatest Common Divisor Deciphered SARS-CoV-2 Superpandemic. Appli Microbiol Open Access. 9:244.
Received: 17-Jan-2023, Manuscript No. AMOA-23-21409; Editor assigned: 19-Jan-2023, Pre QC No. AMOA-23-21409 (PQ); Reviewed: 06-Feb-2023, QC No. AMOA-23-21409; Revised: 14-Feb-2023, Manuscript No. AMOA-23-21409 (R); Published: 22-Feb-2023 , DOI: 10.35248/2471-9315.23.9.244
Copyright: © 2023 Dong KJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.