Journal of Theoretical & Computational Science
Open Access
ISSN: 2376-130X
ISSN: 2376-130X
Research Article - (2016) Volume 3, Issue 2
Herein, we report a one-pot, efficient, and environmentally benign synthesis of biological and pharmacological potent tetrazines and tetrazines were synthesized via reaction of benzyl cyanide and hydrazine in the presence of sodium nitrite and hydrochrloic acid at room temperature in bronsted acidic ionic liquid as a green solvent and efficient catalytic media. Further, computational studies of the synthesized tetrazines have been performed against Human cytomegalovirus (HCMV) protease and found to inhibitors for HCMV. Therefore, tetrazines have been proposed as potent antiviral agents.
<Keywords: Ionic liquid, Tetrazines, One-pot synthesis
Designing of one-pot reactions is a major challenge for the organic and pharmaceutical chemist in academic and industrial world. Such reactions are used for carrying out two or more than two steps reactions in one pot. A huge library of heterocyclic compounds and complicated structures from simple raw materials can be achieved by these methodologies. These processes are very near to an ‘‘ideal organic or inorganic synthesis’’, and via these methodologies, duration of reaction time can be decreased, excellent yields and selectivities can be obtained [1]. Herein no typical equipment/ set-up, expensive protection/ deprotection reagents or steps, purification procedures are required. These methodologies are known as environmentally benign and atom economical processes [2-4].
Tetrazines are heterocyclic compounds and they have attracted the researchers due to their interesting biological activities like intracellular small molecule imaging, genetically targeted protein tagging, post-synthetic DNA labeling, and in-vivo imaging etc. [5,6]. The easy and efficient route for the synthesis of 1,2,4,5-tetrazines can be done by the addition of hydrazine to aromatic cyanide or aromatic nitriles followed by the oxidation of the 1,2-dihydrotetrazine product [7,8]. Although, this methodology has some demerits like it does not work for producing tetrazines from unactivated nitriles means with alkyl nitriles [9-12]. Therefore, developing of easy, efficient and highly selective one-pot processes is urgency in the world of organic synthetic chemistry. Ionic liquids (ILs) have attracted the extensive interest as benign reaction media as well catalyst for carrying the organic reactions due to their unique properties of non-volatility, non-flammability, recyclability, and ability to dissolve a wide range of materials [13-15]. Due to their green credentials and potential to enhance yield, rates of reaction and selectivity, they have been found increasing applications to organic synthesis [16,17].
Human cytomegalovirus (HCMV), a betaherpesvirus, is an opportunistic pathogen that commonly afflicts immuno compromised (HIV) or immunosuppressed (organ/bone marrow transplant) persons. Unfortunately for these patients, current treatments (ganciclovir, foscarnet sodium, cidofovir, and fomivirsen sodium) suffer from cross-resistance, dose-limiting toxic side effects and are only moderately effectual. As a result, in an effort to identify inhibitors with novel mechansims of action, a number of laboratories have sought inhibitors of the essential protease encoded by the HCMV UL80 gene as potentially effective antiviral chemotherapeutic agents. Thus far, several mechanism and peptide-based inhibitors of this enzyme have been disclosed [18-20].
We speculated that the use of bronsted acid ILs might promote this reaction by binding to the nitrile to hydrazine. This is the first report on the bronsted acid ILs, can activate nitriles for nucleophilic addition and promote the formation of 1,2,4,5-tetrazines. Further, they have been used against Human cytomegalovirus (HCMV) protease and proposed them as potent antiviral agents.
Synthesis of Bronsted acid ionic liquid {N-Methyl-2-oxo- N-(3-sulfo-propyl)-pyrrolidinium triflate, MOSPPOTf}
A recyclable bronsted acid ionic liquid was synthesized as in Scheme 1. Equimolar amount of N-methyl-pyrrolidone (20 mmol) and 1,3-propanesultone (20 mmol) were allowed to react in the absence of solvent at 35°C to produce zwitter ionic pyrrolidinium salt in quantitative yield. Further, obtained zwitter ionic pyrrolidoium salt was reacted with an equimolar amount of trifluoromethanesulfonic acid at 135°C with continuous stirring, which yield bronsted acid ionic liquid in quantitative yield.
Optimization of the catalytic system for the synthesis of 3,6-dibenzyl-[1,2,4,5]tetrazine
Different catalytic system has been used to catalyze the synthesis of tetrazines i.e., 3,6-dibenzyl-[1,2,4,5]-tetrazine via the reaction between aromatic/ aliphatic nitriles with hydrazines and the reaction was carried out for appropriate time in the presence of sodium nitrite and hydrochloric acid. Conversion of reactants into product was determined by thin layer chromatography and after the completion of reaction; the compound was extracted with diethyl ether followed by hexane from the reaction mixture. The organic layer was evaporated under vaccume, and yields the corresponding product as in Scheme 2. Structural assignments of the products are based on their 1H-NMR, 13C-NMR, FTIR and mass spectroscopic techniques. Other ionic liquids carrying other counter anions, such as N-Methyl-2-oxo-N-(3-sulfo-propyl)-pyrrolidinium tetrafluoroborate (MOSPPBF4) and N-Methyl-2-oxo-N-(3-sulfo-propyl)- pyrrolidinium hexafluorophosphite (MOSPPPF6), organic solvents like dioxane, acetonitrile, DMF, tolouene and other catalyst were used for the synthesis of 3,6-dibenzyl-[1,2,4,5]tetrazine as in Table 1. We observed maximum yield was obtained in case MOSPPOTf in short span of time at a temperature of 35°C. Similarly, other products i.e., symmetrical and asymmetrical tetrazines were synthesized and characterized as in Table 2.
| S No | Solvent/catalyst | t (h) | Temp. (°C) | Yield (%) |
|---|---|---|---|---|
| 1 | Dioxane | 36 | 35 | 75 |
| 2 | CH3CN | 30 | 35 | 78 |
| 3 | DMF | 32 | 35 | 52 |
| 4 | Toluene | 38 | 35 | 70 |
| 5 | Et3N | 25 | 35 | 45 |
| 6 | None | 36 | 35 | Complex |
| 7 | None | 48 | 75 | Complex |
| 8 | DMSO | 36 | 35 | 50 |
| 9 | THF | 36 | 45 | 60 |
| 10 | MOSPPOTf | 8 | 35 | 94 |
| 11 | CH3CN | 48 | 60 | 80 |
| 12 | Ni(OTf)2 | 24 | 60 | 90 |
| 13 | NiI2 | 24 | 60 | 85 |
Table 1: Optimization of catalytic condition for the synthesis of 3,6-dibenzyl-[1,2,4,5] tetrazine (Entry 3, Table 2).
| C. No. | Analytical data of selected compounds synthesized as in Table 2 | |||
|---|---|---|---|---|
| 1 | 1H NMR (CDCl3) 1.21 (t, 6H, CH3), 1.75 (m, 4H, CH2), 2.76 (q, 4H, CH2) | 13 NMR (CDCl3) Aliphatic carbons (15.2, 27.3, 68.4), aromatic carbon (175.8) | ||
| 2 | 1H NMR (CDCl3) 1.08 (t, 6H, CH3), 1.45 (m, 4H, CH2), 1.80 (m, 4H, CH2), 2.67 (q, 4H, CH2) | 13 NMR (CDCl3) Aliphatic carbons (10.5, 29.5, 37.7, 40.5), aromatic carbon (172.7) | ||
| 3 | 1H NMR (CDCl3) 3.92 (s, 4 H, CH2), 7.08 (m, 10 H, Aromatic) | 13 NMR (CDCl3) Aliphatic carbons (45.5), aromatic carbons (122.2, 125.6, 130.4, 139.6, 168.6) | ||
| 4 | 1H NMR (CDCl3) 3.89 (s, 4 H, CH2), 4.75 (s, 4 H, CH2), 7.12 (m, 8H, Aromatic) | 13 NMR (CDCl3) Aliphatic carbons (40.2, 52.6), aromatic carbons (122.2, 125.6, 130.4, 139.6, 168.6) | ||
| 5 | 1H NMR (CDCl3) 3.95 (s, 4 H, CH2), 6.85 (m, 8H, Aromatic) | 13 NMR (CDCl3) Aliphatic carbons (43.4), aromatic carbons (121.5, 135.1, 139.5, 156.2, 165.2) | ||
| 9 | 1H NMR (CDCl3) 3.91 (s, 4 H, CH2), 7.18 (m, 8H, Aromatic) | 13 NMR (CDCl3) Aliphatic carbons (43.4), aromatic carbons (105.6, 119.1, 132.5, 141.8, 166.8) | ||
| 7 | 1H NMR (CDCl3) 1.12 (t, 6H, CH3), 1.45 (m, 8H, CH2), 1.92 (m, 4H, CH2), 2.50 (q, 4H, CH2) | 13 NMR (CDCl3) Aliphatic carbons (11.5, 25.3, 35.4, 37.2, 40.1), aromatic carbon (169.1) | ||
| 8 | 1H NMR (CDCl3) 2.46 (s, 6H, CH3) | 13 NMR (CDCl3) Aliphatic carbon (21.2), aromatic carbon (164.5) | ||
| 9 | 1H NMR (CDCl3) 1.35 (t, 6H, CH3), 2.69 (t, 4H, CH2) | 13 NMR (CDCl3) Aliphatic carbons (18.2, 28.5), aromatic carbon (168.7) | ||
| 10 | 1H NMR (CDCl3) 3.95 (d of triplet, 4H, CH2), 2.82 (t, 4H, CH2), 1.96 (t, 1H, OH) | 13 NMR (CDCl3) Aliphatic carbons (41.5, 62.8), aromatic carbon (169.2) | ||
| 11 | 1H NMR (CDCl3) 4.85 (d, 4H, CH2), 7.35 (m, 8H, aromatic), 1.98 (t, 1H, OH) | 13 NMR (CDCl3) Aliphatic carbons (69.8), aromatic carbon (125.5, 128.5, 136.8, 141.6, 165.7) | ||
| Reactant 1 | Reactant2 | Product | C. No. | Yield (%) |
 |
 |
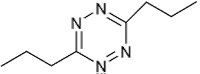 |
1 | 92 |
 |
 |
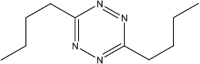 |
2 | 90 |
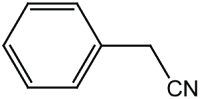 |
 |
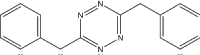 |
3 | 94 |
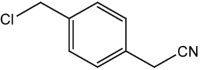 |
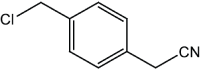 |
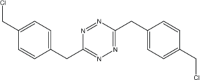 |
4 | 95 |
 |
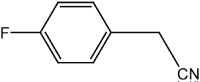 |
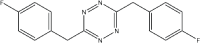 |
5 | 92 |
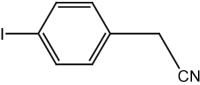 |
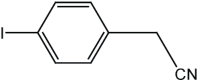 |
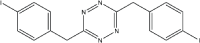 |
6 | 92 |
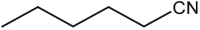 |
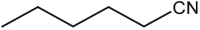 |
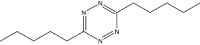 |
7 | 90 |
| CH3CN | CH3CN | 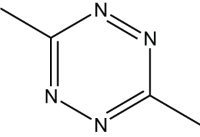 |
8 | 90 |
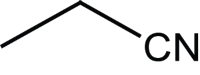
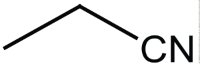 |
 |
 |
9 | 92 |
 |
 |
 |
10 | 96 |
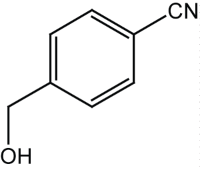 |
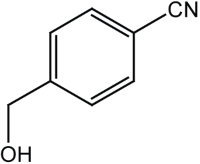 |
 |
11 | 96 |
 |
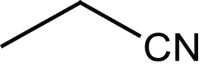 |
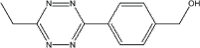 |
12 | 96 |
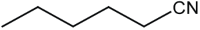 |
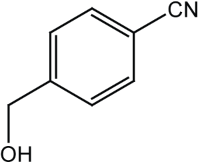 |
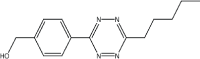 |
13 | 97 |
 |
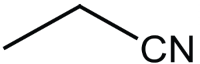 |
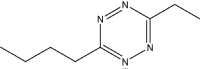 |
14 | 95 |
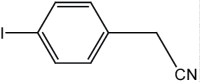 |
 |
 |
15 | 95 |
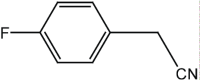 |
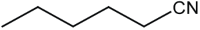 |
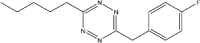 |
16 | 96 |
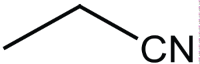 |
 |
 |
17 | 94 |
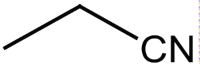 |
 |
 |
18 | 94 |
Synthesis of tetrazines at 35°C using bronsted acid ionic liquid.
Computational study of tetrazines as Human cytomegalovirus (HCMV) protease inhibitors
Optimization of tetrazines: All the tetrazines (18 compounds) were drawn on Chem Draw and were imported to Chem Draw 3D followed by their optimization using MOPAC (a computational tool) suing AM1 calculation and closed shell restricted with RMS gradient 0.1.
Protein preparation: The crystal structure of Human cytomegalovirus (HCMV) protease having PDB ID: 1CMV was taken (http://www.rcsb.org/). The protein was prepared using Molegro Molecular Viewer 2.5 and the same was used for the virtual drug screening.
Virtual screening: Herein, iGEMDock was used for the drug screening as in Table 3. iGEMDock is a molecular docking tool and generates diversity of tetrazines conformations from different seeds with high temperature molecular dynamics. Then, it orients the tetrazines conformations within the defined protein active site by translating the center of the surfactant. Each orientation is subjected to simulated annealing molecular dynamics and sorted according to the interaction energy. iGEMDOCK energy function consists of electrostatic, steric, and hydrogen- bonding potentials. Steric and hydrogen bonding potentials use a linear model. There are four main steps which are used here. Parameters used for drug screening in iGEMDOCK were as followed: initial step sizes (σ=0.8 and ψ=0.2), family competition length (L=2), population size (N=200), and recombination probability (pc=0.3). Optimization is set to generate 70 iterations for which it generated 1200 solutions in one generation process and if exceeded then it terminated after 84,000 solutions. (Yen, Fu 2007) Parameters in iGEMDOCK were set for the successful screening of drug like molecules are as follow: initial step sizes (r=0.8 and w=0.2), family competition length (L=2), population size (N=300), and recombination probability (pc=0.3). Optimization is set to generate 70 iterations for which it generated 1200 solutions in one generation process and if exceeded then it terminated after 84,000 solutions.
| C. No. | Energy | VDW | H-Bond | E (pharma) | H-M-ARG-22 | H-M-GLN-25 | H-M-LEU-32 | V-M-ARG-22 | V-M-TYR-23 | V-M-GLN-25 | V-S-PRO-27 | V-M-LEU-34 | V-S-ARG-166 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | -85.6 | -67.27 | -18.27 | -124.4 | -6.92 | -4.53 | -6.81 | -6.09 | -4.82 | -5.86 | -6.26 | 1.62 | -6.36 |
| 2 | -80.2 | -58.73 | -21.44 | -80.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | -84.3 | -74.99 | -9.30 | -84.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | -98.9 | -73.99 | -24.94 | -98.9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | -87.7 | -79.05 | -8.60 | -87.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | -92.3 | -74.86 | -17.4 | -112.6 | -5.00 | -2.03 | 0 | -4.32 | 0.68 | 1.46 | -6.02 | -5.74 | -5.49 |
| 7 | -88 | -72.08 | -15.89 | -108.2 | -5.63 | 0 | 0 | -4.89 | -4.89 | -1.11 | -5.32 | -2.19 | -4.30 |
| 8 | -70.8 | -49.77 | -21 | -111.4 | -7 | -7 | -7 | -4.50 | -2.30 | -4.55 | -5.26 | -3.40 | -4.79 |
| 9 | -79.4 | -59.78 | -19.60 | -118.2 | -5.60 | -7 | -7 | -5.31 | -2.36 | -4.99 | -4.08 | -6.26 | -4.87 |
| 10 | -89.5 | -62.47 | -27.00 | -128.1 | -5.56 | -6.72 | -7 | -5.27 | -4.39 | -4.98 | -6.25 | -0.93 | -5.63 |
| 11 | -93.2 | -80.43 | -12.75 | -100.7 | 0 | 0 | 0 | 1.46 | -6.24 | -5.36 | -2.98 | -9.59 | -0.11 |
| 12 | -98.6 | -78.07 | -20.54 | -140 | -7 | -7 | -6.54 | -4.73 | -5.82 | -5.04 | -4.56 | -8.14 | -1.97 |
| 13 | -99.3 | -80.78 | -18.49 | -99.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | -89.3 | -68.35 | -20.92 | -130.9 | -6.92 | -7 | -7 | -4.91 | -5.90 | -4.69 | -4.61 | -4.66 | -4.17 |
| 15 | -111.7 | -91.39 | -20.28 | -157.5 | -7 | -7 | -6.28 | -5.04 | -5.24 | -10.35 | -6.21 | -8.47 | -5.75 |
| 16 | -110.3 | -91.79 | -18.52 | -138.2 | -7 | 0 | 0 | -8.41 | -2.05 | -6.48 | -4.02 | -4.48 | -8.11 |
| 17 | -86.8 | -68.87 | -17.95 | -122.8 | -4.01 | -6.93 | -7 | -5.20 | -2.81 | -5.36 | -5.97 | -0.09 | -7.72 |
| 18 | -83.8 | -57.46 | -26.36 | -122.6 | -9.64 | -6.98 | 0 | -6.32 | -4.76 | -4.56 | -1.92 | -6.23 | -3.44 |
Table 3: Docking studies of the synthesized tetrazines as in Table 2.
Herein, a bronsted acidic ionic liquid was synthesized and was used a green and efficient solvent for the synthesis of biological potent symmetrical and asymmetrical tetrazines. A mixture of phenylacetonitrile (nitrile) was reacted in its own in the presence of hydrazine underwent one-pot using MOSPPOTf, a solvent producing compound as in Entry 3, Table 1. The activity of synthesized IL, MOSPPOTf, was compared with other solvent as mentioned in Table 1 in terms of yield and duration of reaction. We observed that the IL, MOSPPOTf showed the best catalytic activity amongst all mentioned in Table 1. Similarly, different tetrazines have been prepared as in Table 2. The advantage of this method is that compounds as in Table 2 were obtained in quantitative yields directly without formation of any byproduct (Scheme 2). Further, docking studies of tetrazines have been carried out using computational tools, iGEMDock. Docking studies are used extensively in drug discovery such as in the prediction of ligandreceptor complex structures and also to rank the ligand molecules based upon the binding energies of the corresponding ligand-enzyme complex. The objective of our docking study is to elucidate the potential interaction mode of the tetrazines with HCMV-protease. The binding energies of the best-docked inhibitor-enzyme complexes are shown in Table 3. A general conclusion derived from these docking results is that the side chain of the Leu 34 forms a hydrogen bond with the inhibitors. The nature of these hydrogen bonds involves an interaction between the amidic nitrogen of HCMV-protease and the nitrogen of tetrazines. Interestingly, this interaction is almost conserved with all studied inhibitors. The docking calculations provided us with a general static picture of the most energetically favourable binding orientation of inhibitors to the enzyme. To obtain further insight into the dynamic changes of the docked inhibitors within the enzyme active site pocket over time, the lowest energy docked complex of the most active inhibitor, 15, was subjected to unconstrained MD simulations and shown in Figures 1 and 2. The post dynamic monitoring of the overall hydrogen bond networks and the electrostatic interaction between the 15 inhibitor and nearby residue clearly revealed that these inhibitors settle comfortably inside the HCMV-protease active site.
In conclusion we have discovered that bronsted acid ionic liquid can be used to catalyze the one-pot synthesis of 1,2,4,5-tetrazines from activated and unactivated nitriles means aliphatic and aromatic nitrile respectively. These tetrazines were not practically accessible and cannot be obtained in high yield even after multistep syntheses, also takes plenty of time. This potential of the process to get a variety of symmetric and asymmetric tetrazines from nitriles can be benefited for the academicians and industrialists. This process will surely be benefited in coordination chemistry, explosives research, materials science, and total synthesis. This process will allow the syntheses of the typical compounds, which were not so easy or able to synthesis in satisfactory yield. Further, computational studies of the synthesized compounds have been performed against Human cytomegalovirus (HCMV) protease and proposed them as potent antiviral agents. Among the all tetrazines, compound no. 15 has shown potent atypical anti-protease activity probably by inhibition of Human cytomegalovirus (HCMV) protease receptors.
Author is highly thankful to Department of Science and Technology (DST), India for their financial support.