Clinical & Experimental Cardiology
Open Access
ISSN: 2155-9880
ISSN: 2155-9880
Protocol Article - (2018) Volume 9, Issue 9
Keywords: Micro-CT Heart; Micro-CT processing; Heart anatomy; Postnatal animal imaging
Three-dimensional (3D) micro-visualizations of biological tissues provide valuable means for morphology study. A wide range of imaging techniques have been described, including confocal microscopy, episcopic microscopy, optical projection tomography (OPT), micro-magnetic resonance imaging (micro-MRI), and microcomputed tomography (micro-CT) [1-6]. Tissue processes for confocal and episcopic microscopy, of which are serial sectional imaging techniques, destroys tissue integrity and prohibits their future use [4,5]. Whole-volume imaging techniques such as OPT, micro-MRI, and micro-CT allow tissue-preservation; however, OPT has restrictions on tissue size [3] while micro-MRI requires prolonged scanning time for maximal resolution of 20 μm/voxel [6]. Micro-CT, on the other hand, has recently attracted great attention from researchers for its highresolution, versatility, availability, and ease of use characteristics [7-11]. Traditionally, micro-CT has limited roles in biomedical studies due to its X-ray attenuation demands from tissue-samples. These limitations, however, have been lifted by the establishment of new staining methods, which showed promising findings in embryology, oncology, histology, angiogenesis, and bioengineering studies [1,8,9,12-19]. Although these studies have established successful staining and micro-CT scanning protocols for small tissues and animal embryos, methods for postnatal animals with developed skeletal systems have not been addressed. Furthermore, while Metscher (2009) reported both iodine and phosphotungstic acid (PTA) are safe and effective contrasts for chicken embryos staining [8], they have not been tested for postnatal animals, cardiac tissues in particular. Furthermore, image quality and anatomical details of the heart shown by in vivo and ex vivo micro-CT scans have not been evaluated against H&E light microscopy. Moreover, effects of CT perimeters and potential denoising algorithms on the image quality of cardiovascular micro-CT should be clarified.
Poor image signal-to-noise indexes have always been a challenge for biomedical image analysis, especially in X-ray dependent imaging modalities. This is often a result of non-uniform X-ray attenuation characteristics associated with different tissues types in a body. It is therefore conceivable that different denoising algorithms have been developed over the years, including median [20-22], anisotropic [23,24], total variation [25-27], and block-matching 3-dimensional (BM3D) [28,29] as attempts to improve image quality. Although each of these techniques has slightly different approaches to image filtering, they were all developed for the ultimate goal of noise removal while preserving image features, particularly edge-enhancements of different objects. We chose to implement the non-local means (NLM) denoising filter in both 2-dimensional (2D) and 3-dimensional (3D) data for its excellent edge preserving property and relative simplicity in application using a modern general-purpose-graphic-processing unit (GPGPU).
In this study, we aim to complement the established tissue preparation and micro-CT scanning protocols to include imaging methods for animal hearts and demonstrate the following:
1. The anatomical details of rats’ hearts revealed by micro-CT are comparable to those of low-field H&E light microscopy.
2. Reduction in the voltage and current of X-ray source increases in vivo micro-CT image degradation. Voltage changes have more detrimental impact than current.
3. Image clarity is proportional to CT scanning time.
4. Iodine is more efficient than PTA in generating uniform staining.
5. Non-uniform contrast staining potentiates image degradation triggered by perimeter changes of X-ray source.
6. Both iodine and PTA-stained heart tissues can be used for subsequent histology processing without compromise.
7. Post-acquisition image processing with NLM filtering can provide modest improvement in image quality, particularly with in vivo micro- CT heart scans.
Compliance with ethical practice
All tissues and animals used in this study were handled with strict compliance to ACT Health Human Research Ethics Committee (ACTH-HREC) and Australian National University Animal Experimentation Ethics Committee (ANU-AEEC).
Iodine and PTA diffusion staining protocols
Thirteen 72 or 96 hrs old neonatal rats were tested. These rats were over anaesthetized with 5% isoflurane and carefully culled via abdominal aortotomy and subsequent thoracotomy. The cardiothoracic tissues were washed with 10% PBS solution for 30 mins to remove residual bloods. Next, the tissues were fixed in 4% formalin solution for 24 hrs until tissue staining.
Two different contrasts were tested based on the promising results of previous study: iodine and PTA [8]. Staining was completed by first subjecting the formalin-fixed tissues to progressive ethanol series to remove formalin: 20%, 50%, 70%, and 90%, each for 1 day respectively. The ethanol-fixed rat’s body was then immersed in 1.5% Iodine (in 90% ethanol), 0.5% PTA (in 70% ethanol), and 1.0% PTA (in 70% ethanol) for various periods prior to scanning as listed in Table 1.
| Rat heart, (Rat age) | Stain type | Stain concentration | Effective Staining duration | Micro-CT type | Scanning time | Resolution (µm) | Voltage (kV) | Current (uA) | Results: Excellent-Detailed anatomy Good-Gross anatomy, sharp border Poor-Gross anatomy, blurry border. Incomplete-Undifferentiated anatomy |
|---|---|---|---|---|---|---|---|---|---|
| 72 hrs | Iodine | 1.5% in 90% Ethanol | 48 days | Ex vivo | 15 hrs | 10.7 | 100 | 100 | Excellent-Figure 2b and 2c |
| 72 hrs | Iodine | 1.5% in 90% Ethanol | 16 days | In vivo | 3 min | 10 | 90 | 200 | Good-Figure 3a |
| 72 hrs | 16 days | 3 min | 70 | 200 | Good-Figure 3b | ||||
| 72 hrs | 16 days | 3 min | 50 | 200 | Poor-Figure 3c | ||||
| 72 hrs | 16 days | 3 min | 30 | 200 | Incomplete | ||||
| 72 hrs | 16 days | 3 min | 90 | 100 | Good-Figure 3d | ||||
| 72 hrs | Iodine | 1.5% in 90% Ethanol | 16 days | In vivo | 3min | 20 | 90 | 200 | Good-Figure 4a |
| 72 hrs | 16 days | 3 min | 70 | 200 | Good-Figure 4b | ||||
| 72 hrs | 16 days | 3 min | 50 | 200 | Poor-Figure 4c | ||||
| 72 hrs | 16 days | 3 min | 30 | 200 | Incomplete-Figure 4d | ||||
| 72 hrs | 16 days | 3 min | 90 | 100 | Good-Figure 4f | ||||
| 72 hrs | 16 days | 26 s | 90 | 200 | Poor-Figure 4e | ||||
| 96 hrs | PTA | 1.0% in 70% Ethanol | 148 days | In vivo | 3 min | 20 | 90 | 200 | Good-Figure 5a |
| 96 hrs | 148 days | 26 s | 90 | 200 | Poor-Figure 5b | ||||
| 96 hrs | PTA | 0.5% in 70% Ethanol | 699 days | In vivo | 3 min | 20 | 90 | 200 | Incomplete-Figure 6a |
| 699 days | 3 min | 70 | 200 | Incomplete-Figure 6b | |||||
Table 1: Summary of various tissue staining and micro-CT setups for different rat hearts with respective image results.
in vivo and ex vivo micro-CT scanning
Current micro-CT systems are generally classified into in vivo and ex vivo based on the system setups; these terminologies are not related to their standard definitions in biomedical science but rather as descriptions of micro-CT system setups [30]. in vivo micro-CT scanner incorporates a stationary sample positioned in between a rotational system of X-ray source and detector. On the contrary, ex vivo micro-CT system involves a rotational sample situated in between an adjustable X-ray source and detectors; this setup can yield a more magnified image by shortening the distance between the sample and X-ray source [31,32]. Furthermore, it can generate a high signal-tonoise ratio image through prolonging the scanning time while keeping X-ray source current low. One of the advantage of keeping low X-ray source low is to minimize X-ray source spot-size and thereby enabling higher visual resolution, <2 μm/voxel [33].
In this study, a commercially available Caliper Quantum FX machine has been chosen to represent the in vivo micro-CT scanners and a custom-built micro-CT system by Applied Mathematical Department of Australian National University (ANU) was used to represent ex vivo micro-CT systems.
Using the Caliper Quantum FX, a tissue specimen is placed on a stationery loading dock positioned in between the rotating system of X-ray source and detector. The scanning time is pre-set between 27 s and 4.5 min depending on the field of view (FOV; ranging from 5-75 mm in diameter) and the chosen modes of quality (ranging from standard to fine). The maximum resolution achievable is 10 μm/voxel. In this study, different combinations of scanning perimeters including scanning time, voltage, and current adjustments were explored to find their effects on imaging quality, as summarized in Table 1. The resultant images were stored as DICOM series and visualized with FIJI and Drishti, both of which are open-source software. Semi-automatic organ segmentation was performed using Drishti paint, a function of Drishti [34,35].
The custom-built micro-CT system by ANU Applied Mathematical Department requires the sample to be fixed within an aluminium tube. This tissue-tube is then placed on a rotational stage situated inbetween an adjustable X-ray source and scintillator CCD. X-ray source and sample distance is adjusted according to the resolution desired [33]. All samples were allocated with 15 h of scanning time and an additional 8 h of image processing and reconstruction time via National Computational Infrastructure (NCI) services. The maximum resolution is 1 μm/voxel, limited by the physical size of the sample. The resultant images were stored as net CDF files and visualized with Drishti [34].
H&E stained microscopy
Following micro-CT scanning of the hearts, the tissues were processed with standard H&E protocols for light microscopy [36].
H&E processing were conducted in the following steps. Firstly, the iodine-stained hearts were sectioned sagittally into 4 mm-thickness blocks and placed in cassettes. Next, these tissue-blocks underwent contrast washout and dehydration in 90% EtOH for 48 hrs. Xylol was then used to clear alcohol before paraffin-embedment at 60°C. Afterward, the tissue samples were sliced into tissue-sheets of 10 μm in thickness using a microtome. These tissue-sheets were laid in waterbath of 5-6°C to reduce wrinkles while being positioned onto the labeled-glass slides. Dehydration was then completed overnight at 37°C. Finally, the coverslip was fixed to the slide using Eukitt® resinous medium.
Progressive H&E staining was adopted. The slides were first stained in alum-haematoxylin solutions until the appearance of dark red colour; this was followed by washing and ‘bluing’ with lithium carbonate solution. Finally, the slides were then washed followed by counter-stained with 0.5% eosin alcoholic solution.
All tissue slides were examined using Olympus IX 71 microscope.
Post-acquisition image processing: Non-Local Means (NLM) filtering
As an attempt to improve image clarity and edge-sharpness of cardiovascular micro-CT scan, we trialed NLM filtering using customcode.
NLM algorithm was first developed by Buades et al. [37] under the premises that similarity exists among different regions of an image provided that these regional sizes are 'relatively' small. This selfsimilarity enables noise cancelling by averaging these similar parts of the image. Genin et al. (2012) has illustrated the computational principle of this algorithm [38]. In 2D images, this process creates a cube; whereas in 3D images, this generates a 4-dimensional (4D) hyperplane. This now gives the most similar pixels/voxels in the image and the denoising or smoothing operation is done based on this higher dimensional object. This operation is feature-preserving and enabling automated noise removal. This NLM algorithm is widely considered as a state-of-the-art denoising filter. The large number of computations required to perform the basic NLM, however, making its computational-processing-unit (CPU) runtime unacceptably high for all but the smallest 2D images (Figure 1).
Figure 1: Illustration of region-matching of high similar measures in NLM processing. A pixel/voxel is chosen, a voxel being the smallest unit of a 3D volume, analogous to a pixel in 2D image. A neighbouring region with high similar measures around this selected pixel/voxel is then chosen. This neighbouring-region is a square in 2D and a cube in 3D. A sum of the squared difference is used to determine which neighbourhoods are the most similar to the current region of interest; this is then ranked and put into a higher dimensional stacks. Various groups of high-association groups of images are selected based on their similarity measures to each selected reference patches, highlighted in thick black boxes [37].
The method utilized in this paper uses standard NLM but with locally adaptive estimates of noise, which results in more accurate removal of noise. Given all images in this study were all 3D data; 3D cubes were used for the similarity measure and therefore resulting in the use of the 4D hyperplane. Images presented in this paper ranging from 5123 voxels for the in vivo micro-CT scans to the full 20483 voxels for the ex vivo micro-CT scans. Using our proposed protocol (unpublished), the computing time for the NLM has reduced to approximately one-minute-thirty-seconds for a 5123 sample using a single graphics-processing-unit (GPU) card, significantly faster than current commercial vendors with processing time of three hours for a similar sample size.
A brief discussion of NLM implementation used in this study is described as follow. Traditionally, a great majority of the NLM runtime is spent on the computations of the sum of squared differences (ssd). Given the ssd computation for adjacent voxels is highly similar, we now exploit this similarity and adopt a pseudo moving average filter (MAF) to perform the ssd calculations from one pixel/voxel to the next. For 2D images, the squared area of intensities used for one voxel’s ssd-search-neighborhood differs from an adjacent voxel’s ssd-searchneighborhood only in the end rows or columns. The same principle also applies in 3D data; with adjacent cubes differ only in two end slices. To illustrate further, if M is the number of voxels of the inner ssd cube and N is the number of cubes requiring ssd calculations, then the total number of difference-operations is as follows: with no MAF, we performed N × M3 operations; with the MAF, we performed M3+(N-1) × 2 × M2 operations. Based on this, we can expect a significant computational reduction when N is large. This was indeed the case when handing presented ex vivo micro-CT data, where each dataset having an average of 4.7 GB in size, the second operation became at least a two-orders of magnitude less in computations.
The computation of the smoothing parameter and estimation of the image noise is adapted from Coupe et al. [39]. However, our implementations is unique in the fact that we compute individual noise estimates for each sub blocks of the image rather than assigning a single estimate for the entire image. Furthermore, the gray-scale-value (gsv) intensity range has also been taken into account in our computations of the smoothing parameter. The gsv inclusion was found to be empirically important for the success of de-noising a range of image types including computed tomography (CT), scanning electron microscopy (SEM), and ultrasound etc. Moreover, our implementation of NLM follows the original model of Additive Gaussian White Noise (AGWN); however, it also allows adjustment on the shape of the Gaussian kernel to better approximate the real noise distribution for images with non-AGWN.
Our empirical testing confirms that through local estimation of the noise, inclusion of correction factor for non-AGWN, and fixed window sizes, improved de-noised quality can be achieved. Little, if any, gains in quality improvement was obtained with larger search window sizes. On the contrary, image quality may actually degrade with larger search window sizes. These conclusions are drawn based on qualitative and quantitative measures such as peak signal to noise ratio (PSNR), line tests, and residual images for a wide range of image types. Similar finding are also found in Coupe et al. [39].
The implementation command was run on a Linux operating system on computer fitted with Intel (R) Core ™ i7-4770K CPU @3.5 GHz, 32 GB of RAM, and a NVidia GeForce GTX Titan Black Kepler GK110 architecture. The processed CT data were reviewed using Fiji and Drishti [34,35].
Image quality: Comparing light microscopy, ex vivo, and in vivo micro-CT scans of cardiovascular tissues of postnatal rats
To demonstrate the visualization power and versatility of the proposed method for cardiovascular tissue, we compared H&E light micrograph with ex vivo and in vivo micro-CT scans of the same heart, as shown in Figure 2. Although good tissue contrast is observed throughout Figures 2b-2e, only ex vivo micro-CT scans (Figures 2b and 2c) provide good visualization of anatomical details comparable to those seen on 4X H&E light micrographs (Figure 2a). Furthermore, tissue preservation is observed only in micro-CT scans as opposed to apparent micro-tears shown by H&E light micrograph. Moreover, Figures 2b-2e illustrated the versatility and ease of manipulations through different visual planes of the heart using micro-CT data; this was difficult to achieve with light-microscopy. Additionally, when setting both ex vivo and in vivo cardiac micro-CT scans with similar field of view for comparison, the resolution limitations of in vivo micro-CT scans (Figures 2d and 2e), are obvious when comparing to ex vivo micro-CT scans (Figures 2b and 2c). The respective resolutions of in vivo micro-CT and ex vivo micro-CT scans are 40 μm/voxel and 10.7 μm/voxel. In fact, only macroscopic anatomical details such as atria, ventricles, papillary muscles, and even aortic semilunar valves can be seen with in vivo micro-CT scans, whereas small structures including small coronary arteries and even cardiac tissue striations can be easily appreciated with ex vivo micro-CT scans. Overall, in vivo micro-CT scans offered less image clarity and cardiovascular details than those of ex vivo micro-CT scans and H&E light microscopy.
Figure 2: Comparison of H&E microscopy, ex vivo and in vivo micro-CT scans of neonatal rat heart. Figure 2a (H&E microscopy, 4X magnification): 2D view of major heart structures as labelled. Muscle fibre and micro-tears at the base of interventricular septum caused by tissue processing are also illustrated. Figures 2b and 2c (ex vivo micro-CT, resolution 10.7 μm/voxel): structurally intact 3D visualization of heart structures with details comparable to that of H&E light micrograph. Figure 2a. In addition, cross sections of coronary artery and vessel wall thickness can be appreciated. Figures 2d and 2e (in vivo micro-CT, resolution 40 μm/voxel): saggital and parasaggital view of in vivo micro-CT heart scans showing gross but limited macroscopic 3D visualization of heart structures. Aor: Aorta; Rau: Right Auricle; RA: Right Atrium; RV: Right Ventricle; RVW: Right Ventricular Wall; IVS: Interventricular Septum; LA: Left Atrium; LV: Left Ventricle; LVW: Left Ventricular Wall; Pm: Papillary Muscle; Pul V: Pulmonary Vessel; Asv: Aortic Semilunar Valve.
Effect of X-ray source voltage, current and CT scanning time on low resolution (20 μm/voxel) and high resolution (10 μm/ voxel) in vivo micro-CT heart scans.
Since in vivo micro-CT has less-than-ideal image quality, we explored potential methods for improvement through CT-perimeter adjustments. We first tested on low-resolution CT-scans (20 μm/voxel) derived from an iodine-stained heart. As shown by Figures 3a-3d, gradual reductions in X-ray source voltage result in progressive worsening of image clarity. Minor image degradation is observed with voltage reduction from 90 kV to 70 kV; the overall heart structures remain discernible. On the other hand, non-meaningful image is generated when setting X-ray source voltage to 30 kV (Figure 3d). Furthermore, shortening the CT scanning from 3 min to 26 s results in significantly blurrier images exemplified by the compromised pulmonary and aortic vessel definitions (Figure 3e). Unexpectedly, halving the X-ray source current only modestly reduces the sharpness of the image (Figure 3f).
Figure 3: Effect of micro-CT voltage, current, and scanning time on low-resolution (20μm/voxel) in vivo micro-CT images of rat heart. Figure 3a (90 kV), 3b (70 kV), 3c (50 kV), and 3d (30 kV): demonstrate lowering voltage setting results in blurrier structural border, which is best appreciated by the gradual loss of right ventricular papillary muscle outlining as shown by 3a to 3d. Furthermore, minimal anatomical information can be detained from Figure 3d. Figure 3a (3 min) and 3e (26 s): Short CT scanning time generates less homogenous cardiac scan with blurrier edges. Figure 3a (200 μA) and 3f (100 μA): reducing x-ray source current yields compromised cardiac image with lower tissue-cavity contrast.
To investigate further, we tested the effect of similar perimeter adjustments on high-resolution micro-CT scans (10 μm/voxel) using the same iodine-stained heart. Consistent with low-resolution image series, voltage reduction corresponds with poorer image quality (Figures 4a-4c); however, image degradation associated with reduction in X-ray source voltage or current is more prominent in high resolution images. Indeed, direct comparison of Figure 4c with Figure 3c, both derived from an X-ray source voltage setting of 50 kV, shows significant higher image noise and blurrier cardiac edges in 10 μm/ voxel heart scans. Interestingly, halving the current setting from 200 μA to 100 μA (Figure 4d), yielded similar image quality as Figure 4b, a scan generated by 70 kV X-ray source, which is only a 22% reduction from a standard 90kV setting (Figure 4a).
Figure 4: Effect of micro-CT voltage and current on high-resolution (10 μm/voxel) in vivo micro-CT images of rat heart. Figure 4a (90 kV), 4b (70 kV), and 4c (50 kV): longitudinal views of micro-CT heart scans showing reduction of X-ray source voltage results in higher image noise. Image clarity gradually reduced from Figure 4a to 4c as demonstrated by less demarcated ventricular borders. However, macroscopic information including heart chamber dimensions, ventricular wall thickness, and outlining of papillary muscles are preserved, which is best illustrated in left ventricle. Figure 4a (200 μA) and 4d (100 μA): demonstrates halving the micro-CT current setting results in higher image noise. Internal ventricular border shown by Figure 4d is blurrier than that shown by Figure 4a.
Effect of CT scanning time on micro-CT scans of fullystained PTA (1.0%) hearts
PTA staining was tested for the search of alternative contrast to iodine. High radiopaque micro-CT scans were generated with a fully PTA-stained heart (Figure 5). However, a prolonged staining period of 148 days was required to guarantee successful diffusion staining of a neonatal rat’s heart using 1.0% PTA. Furthermore, although PTA staining yields good soft-tissue contrast, its high radiopacity reduces the visibility of structural details, such as loss of vessel boundary between aorta and pulmonary artery or indistinguishable borders between right auricle and ventricle (Figure 5a). Furthermore, shortening CT scanning time causes significant higher image degradation with PTA-stained heart (Figure 5b) than that of iodine stained heart (Figure 3e).
Figure 5: Effects of PTA staining and scanning time on micro-CT scans of neonatal rat heart-148 staining days. Figure 5a (1.0% PTA, 3 min): presents micro-CT scans of a fully PTA-stained heart with strong cardiac parenchyma and cavities contrast. However, due to the strong radio opacity, adjacent structures such as pulmonary and aortic vessel walls cannot be differentiated. Figure 5b (1.0% PTA, 26 s): shorter scanning time CT scanning time gives poor structural details due to higher image noises.
Effect of X-ray source voltage on micro-CT scans of nonhomogenous- stained PTA (0.5%) hearts
To investigate whether high radiopacity of PTA-stained cardiac scan can be corrected by reduction in contrast concentration, we applied diffusion staining with 0.5% PTA contrast for 699 days but with little staining success. As shown in Figure 6a, peripheral cardiac tissue stained by PTA exhibited significant high opacity while internal structures cannot be delineated due to incomplete staining. Furthermore, non-homogenous PTA staining amplifies the image degradation on micro-CT heart scan from lower X-ray voltage settings as shown in Figure 6b.
Figure 6: The effect of non-uniform staining on cardiac tissue micro-CT image-699 days of staining. Figure 6a (0.5% PTA, 90 kV) shows moderate contrast details at the outer rims of the heart with limited internal clarity. Internal structures such as interventricular septum are poorly defined. Figure 6b (0.5% PTA, 70 kV) shows rough outer boundary of stained regions, mainly the outer rims of the ventricles. The boundary of internal ventricular wall and septum cannot be delineated.
Effect of NLM denoising algorithms on ex vivo and in vivo micro-CT rat heart scans
As attempt to improve image quality, we applied the NLM denoising algorithm to both ex vivo and in vivo micro-CT scans for structural feature enhancements and image noise removal. Due to the high signal-to-noise ratio of the original ex vivo micro-CT scan (Figure 7a) only subtle improvement such as boundary differentiation can be appreciated post-filtering (Figure 7b). The subtle improvement is better appreciated by comparing their respective magnified views of intraventricular edges. Furthermore, through NLM filtering, structural boundaries demonstrated in vivo micro-CT scans have become modestly more defined (Figure 7d) when comparing to the original scan (Figure 7c). This can be appreciated by reviewing the aortic and intraventricular edges. In addition, the comparison of their respective intensity histogram also demonstrates significant narrowing of histogram spread.
Figure 7: Cardiac anatomical differentiation of both ex vivo and in vivo micro-CT scans are enhanced by NLM filtering. The original (7a) and denoised (7b) ex vivo scans illustrate that mildly enhanced cardiovascular parenchymal edges in denoised (7b) while anatomical information is unaltered in both. Similar but slightly more prominent enhancements are in 7c and 7d, original and denoised in vivo micro-CT scans, respectively. The respective contrast histograms of each scans revealed narrower histogram peaks in denoised group, 7b and 7d, in comparison to their respective originals, 7a and 7c.
Our study demonstrated micro-CT scanners, with proper tissue staining, can generate scans illustrating detailed cardiovascular anatomy of postnatal rats. These anatomical details revealed by the ex vivo micro-CT scans not only share similar soft-tissue differentiation as micro-MRI scans but also offer high-resolution structural details comparable to low-magnification H&E light microscopy (Figure 2a) [40]. This easy, versatile, tissue-preserving methodology offers a potential alternative method to conventional pathology and morphology study [41,42]. In addition, the benefits of 3D rendering and the ability to accommodate relatively large sample size allow for better internal anatomical visualization and appreciation of disease processes. Although adopting micro-CT modality into clinical practices may still be years away, micro-CT has already taken important research roles using the appropriate preparations and should be introduced to experimental cardiac study.
As previously expected, we confirmed ex vivo micro-CT scanners offer significantly more detailed and clearer images than in vivo micro- CT scanners on imaging cardiovascular tissues (Figure 2). Figures 2b and 2c demonstrate ex vivo micro-CT scans can provide well-defined illustrations of cardiac anatomy in addition to cardiac parenchymal distribution and muscle striations at a level of details comparable to those of 4X H&E light micrograph (Figure 2a). On the contrary, this is not achievable by in vivo micro-CT scans (Figure 4a) despite similar high-resolution settings, 10 μm/voxel (in vivo micro-CT scan) and 10.7 μm/voxel (ex vivo micro-CT scan). The difference in image quality is due to the intrinsically high signal-to-noise ratio achieved by the extended scanning time of the ex vivo micro-CT scanner, i.e. 15 h, comparing to the relatively short scanning period of the in vivo micro- CT scan, 3 mins. The limited scanning time of the in vivo micro-CT scanner is designed to minimize radiation exposure in view of potential live animal study; however, this has significantly reduced physical raw data acquired for downstream image reconstruction, resulting in lower signal-to-noise index and therefore compromised the image quality.
Due to the wide availability and lower cost of in vivo micro-CT scanners, we explored the effects of various settings of the X-ray source’s voltage, current, and CT scanning time on low-resolution and high-resolution scan-setting with the aims to derive the best image quality. Although the full extent of electromagnetism and photoelectric effect of X-ray physics is beyond the scope of this study, for practical purposes of manipulating the settings of in vivo micro-CT scanners, we found our scan result is consistent with the following:
Image clarity ∝ CT Scanning time*Xray Current*Xray Voltage
We confirmed image signal-to-noise ratio can be improved by increasing the current of X-ray source thereby generating of higher number of photons within a short period of time; this benefit is illustrated by the comparison between Figures 3a and 3f. Consistently, image degradation associated with lower current setting is more prominent with high-resolution images, as the comparison showed between Figures 4a and 4d. Moreover, reduced image quality from lower current setting can be rescued by longer scanning time, as demonstrated by the comparison of Figures 3a and 3e.
Furthermore, we confirmed voltage has an important effect on both image acquisition and signal-to-noise ratio. As majority of the ionizing electromagnetic radiation, X-ray, is generated through the Bremsstrahlung process, radiation emission from the “braking of electrons,” a continuous spectrum of the X-ray energy is generated based on the anode voltage of X-ray tube. As shown in Figure 3d, when setting the anode voltage to <50 kV, no meaningful image can be generated from our in vivo micro-CT scan trials. This can be explained by the fact that wide-angle Compton scattering, prominent Rayleigh scattering, and less photoelectric effect predominate at low voltage setting, resulting in less X-ray production [43]. On the other hand, high-voltage setting minimizes Rayleigh scattering and increases the narrow-angle Compton scattering and photoelectric effect thus creating sharper-edge image through X-ray spectrum changes: generating more narrow-angled scattering photons with higher penetration power. However, this change also increases the risk of “over-exposure” [44]. This effect can be seen promptly in high-spatialresolution images (Figures 4b and 4c) and less prominent in lowspatial- resolution scans (Figures 3b-3d). Expectedly, image noises from suboptimal voltage or current setting are accentuated by inadequate staining, as shown by Figure 6b, likely due to uneven photon scattering resulted from non-homogenous distribution of staining effect.
Based on our results, we suggest titrating up the X-ray source current of in vivo micro-CT scanner before altering the voltage for image quality improvement while minimizing the risk of overexposure. On the other hand, when the desired resolution is <2 μm/voxel, an upper limitation of current size exists due to the finite size of generated photon beams, which can be greater than 2 μm and hence unable to project the fine differentiations of the objects. However, to our best understanding, no commercial in vivo scanners offer spatial resolution higher than 10 μm/voxel currently.
From this study, we illustrated that intact cardiovascular tissues can be visualized using micro-CT scanners following iodine or PTA tissuestaining, provided that prolonged PTA staining time can be allocated. Contrary to previous study on animal embryos, diffusion staining on postnatal rat hearts requires much longer staining time to guarantee success, up to 16 days using 1.5% iodine solution and 148 days with 1.0% PTA [8,16]. Staining with 0.5% PTA for 699 days was not successful. The difference in the required staining time was likely due to the size and polarity differences, with iodine being non-polar and about 20 times smaller than the ionic PTA [45,46]. In addition, due to PTA’s high molecular size and higher mass absorption coefficient, we found attempting to acquire high-resolution images (i.e. 10 μm/voxel) using in vivo micro-CT scanner runs a higher risk of generating “overattenuated” image (Figure 6a) [47]. This observation is consistent with Lambert’s law [48,49]:
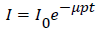
Where, =resultant intensity, =initial intensity, =mass absorption coefficient (cm2/g), =density (g/cm3), =thickness (cm).
Furthermore, to explore rescuing measures for suboptimal cardiac micro-CT images, we implemented NLM algorithms for image processing; a process that was known to be time-consuming until parallel processing was adopted using GPGPU. We found high signalto- noise ex vivo micro-CT scans exhibit only minor improvements from such processing, as shown in Figures 7a and 7b. These improvements were slightly more prominent with low signal-to-noise in vivo micro-CT scans, as shown in Figures 7c and 7d. By adopting this process, further quantitative analysis may be improved from clearer images. While our implementation is still in trial phase and not available for public domain; other alternative open-source-projects are available for NLM implementation, including Fiji, albeit the processing may be slower with more artefact in resultant images [35]. Although improvement from NLM may be minor currently, de-noising can facilitate future development of automated segmentation and hence accurate automated quantitative analysis such as measurement of organ sizes.
In conclusion, we demonstrated that micro-CT is a tissuepreserving scanning modality able to provide detailed cardiovascular anatomy with great tissue differentiation using simple diffusion staining methods. This potentially opens up different applications for micro-CT scans. Contrary to prior findings from embryo study, we recommend iodine as the preferred contrast for efficient and reliable results. Ideally, we recommend ex vivo over in vivo micro-CT scanner for achieving high-definition heart images. However, if an in vivo micro-CT set up needs to be used, we recommend increasing current and/or CT scanning time before altering voltage towards extreme setting for clearer image acquisition. Lastly, despite of macroscopic coloration by iodine staining, we have confirmed that it does not interfere with further H&E processing on the tissue. Through the same process, we also illustrated ex vivo micro-CT scans can offer similar cardiac details as low-power microscopy, as shown in Figures 2a-2c. Furthermore, quantitative study of cardiac fibrosis post-myocardial infarction can be possible through measurement of the loss of cardiac muscle striation using volume rendering software, such as Drishti [34]. Similarly, quantitative analysis on the gross anatomy of aortic dimensions, ventricular volumes, valvular sizes, and muscular hypertrophy can be completed using micro-CT data. Lastly, in consideration of the efficiency of fast NLM de-noising and suboptimal images of in vivo micro-CT scans, we recommend in vivo micro-CT images to be processed by NLM algorithms prior quantitative analysis.