Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2012) Volume 5, Issue 2
Chikungunya is viral disease is transmitted to human beings by mosquito bite. Chikungunya can cause fever, chills, nausea, vomiting, joint pain, head ache, and swollen in the joints. A cause of Chikungunya is mainly due to the replication of Chikungunya virus in the human body. The nsP2 (non structural Protein 2) protein is responsible for Chikungunya virus replication. CHIKV replication causes general host shut-off. CHIKV replication is resistant to inhibition by interferon once RNA replication has been established and that CHIKV actively suppress the antiviral IFN response by preventing IFN-induced gene expression. Chikungunya virus replication and propagation is dependent on the protease activity of the viral nsP2 protein, which cleaves the nsP1234 polyprotein replication complex into functional unit This study investigates about the nsP2 protease protein of Chikungunya 37997 and computational methods have been used to find the best interaction site in the protease protein and identify the best ligand compounds for inhibiting the nsP2 protein. The Homology Modelling was used to predict the 3-Dimensional structure of nsP2 Protease protein. The 3D structure was validated using Structure Validation Server (SAVS) in which 85.2% of residues present in the favored regions of the Ramachandran Plot. Docking studies were carried out with various inhibitors and it was found that LIGAND_4 ( N-butyl-9-[3,4-dipropoxy-5-(propoxymethyl)oxolan-2-yl] purin-6-amine ) had the most stable interaction with the nsP2 CHIKV Protease. Indicate that LIGAND_4 could be a promising inhibitor for nsP2 Protease of CHIKV virus as the drug target.
Bioinformatics, combination of biotechnology and computer science plays on important role in the design of new drug compounds [1]. Structure based drug design is perhaps the most elegant approach for discovering compounds exhibiting high specificity and efficacy and is also important because of its impact on the economics of drug discovery [2]. Chikungunya is a viral disease that is transmitted to human beings by mosquito bite of Aedes egypti. It can cause joint pain, chills, nausea, vomiting, fever, and head ache [3]. The cause of Chikungunya is mainly due to the replication of CHIKV virus inside the human body. If Forest day mosquito bites an infected person, Then the virus gets consign to its body. The virus does not infect the mosquito itself, but on the adverse, the mosquito bites a healthy person, the enhanced virus is now transferred into the body of the healthy person. The Chikungunya virus depends upon the process of viral tropism for its action. Once the virus enters into a host it searches for susceptible cells. As soon as it finds one, the virus fuses with the cell membrane of the cell and it proceeds towards the nucleus of the cell. It then enters the nucleus and hence captures the biosynthetic machinery of the cell, and directs the synthesis of viral proteins as well as the replication of the viral genome. The mode of action of Chikungunya virus, by which it causes the disease remain to be investigated in detail and its mechanism of action has not yet been fully characterized accept the fact that it causes major histopathological changes in the skeletal muscle tissue, severe inflammation and necrosis of skeletal muscle.
The non structural proteins play an important role in CHIKV viral replication, newly synthesized non structural proteins from the replication that mediates the synthesis of viral RNA’s. This CHIKV replication causes general host shut off. This will leads to severe cytopathicity in mammalian cells. Once RNA replication has been established, CHIKV replication is resistant to inhibition by interferon. The CHIKV actually suppress the antiviral IFN response by preventing the IFN induced gene expression.Chikungunya virus replication and propagation mainly depends on the protease activity of the viral nsP2 protein, which cleaves the viral nsP1234 polyprotein replication complex into functional unit. There is evidence that this protease protein plays some role in the replication of virus particle in the human beings, Therefore nsP2 protease is responsible for cleavages in the non structural polyprotein that are important for the viral replication cycle. Therefore this protease constitutes an attractive drug target for the development of antiviral compounds. Our work is mainly focused on the interaction of protease protein with the potent novel inhibitor. These ligands were screened form PubChem database by ADMET Properties that is aqueous solubility, Blood Brain Barrier penetration, Hepatotoxicity, Human Intestinal absorption and Plasma Protein Binding and Drug likeliness properties.
The protein molecule selected for the docking studies was chikungunya nsP2 Protease.
Ligand selection
PubChem Database compound contains validated chemical depiction information. Structures that are stored in PubChem contain calculated properties and description which helps in searching and filtering of chemical structures. The ligand for nsP2 protease protein was retrieved from PubChem Compound. The ligand structure, name and molecular formula are given in Table 2. The ligands were downloaded as XML file format from PubChem compound. The XML files were converted into the 3D structure using Open Babel software.
| S.No. | Name | Description |
|---|---|---|
| 1. | Accession | AAU43880 |
| 2. | UniProt | Q5XXP4 |
| 3. | Taxonomic Identifier | 371095 |
| 4. | Protein Name | Non-Structural Protein 2 |
| 5. | Virus Host | Aedes aegypti |
| 6. | Organism | Chikungunya virus (strain 37997) (CHIKV) |
Table 1: Shows the information about the Target Protein CHIKV nsP2 Protease Protein.
| S.No. | Structure | Name of the Lead | Molecular Formula |
|---|---|---|---|
| Ligand_1 | 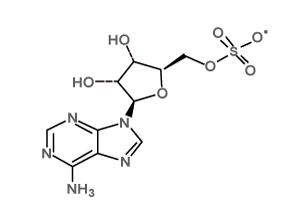 |
[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl sulfate; tetrabutylazanium |
C26H48N6O7S |
| Ligand_2 | 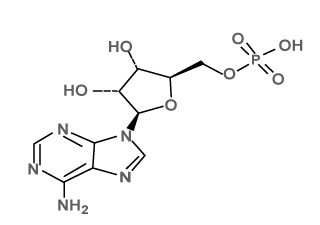 |
[5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl hydrogen phosphate; tributylazanium |
C22H41N6O7P |
| Ligand_3 | 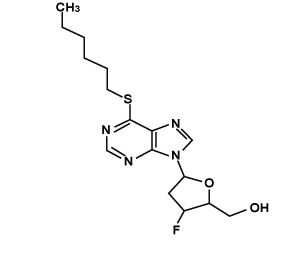 |
[3-fluoro-5-(6-hexylsulfanylpurin-9-yl)oxolan-2-yl]methanol | C16H23FN4O2S |
| Ligand_4 | 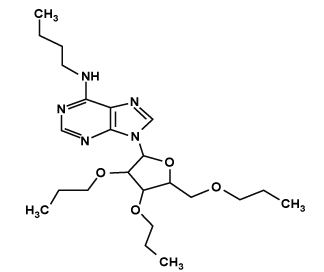 |
N-butyl-9-[3,4-dipropoxy-5-(propoxymethyl)oxolan-2-yl]purin-6-amine | C23H39N5O4 |
Table 2: Screened Ligand Molecules form PubChem Database.
Dataset and 2d similarity search
The chemical structure of the Protease inhibitor such as S-adenosyl methionine was collected from published literature with their biological activity data. In this study, 2D analogues of the nsP2 protease inhibitors were retrieved from the PubChem database using PubChem Structure search tool by means of the 2Dsmiles of the inhibitors. In this similarity search, compounds with desired physicochemical property were retrieved using the threshold values for Drug likeliness and exported in SDF format [8].
SMART screening
DS-smarts screening filter was used to screen the screened ligands from the library of the compounds using the Discovery studio2.5. Discovery studio package contains the smiles of the Reactive functional group of the database and the compounds screened using the DS_ smarts [8]. Usually many drugs fail in clinical trials because of unrelated side effects and Bio-Unavailability. To overcome these problems, we screened drug like molecules that exhibit physicochemical properties for favorable absorption, distribution, metabolism, excretion and toxicological parameters. Key physicochemical properties of compounds such as Molecular weight, ClogP, PSA, Heavy atoms, HBA (Hydrogen Bond Acceptor), HBD (Hydrogen Bond donor), Rotatable bonds, and number of Rings were considered in this analysis.
ADMET
Discovery Studio provides methods for assessing the disposition and potential toxicity of a ligand within an organism. The ADMET protocols contain published models that are used to compute and analyze Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties [8]. In addition, specific rules were applied to remove ligands that are not likely drug-like, unsuitable leads, etc. based on the presence or absence and frequency of certain chemical groups.
The ADMET functionality test was used to estimate the aqueous solubility of a set of ligands, estimate the Blood brain barrier penetration (BBB), estimate Cytochrome P450 (CYP450) 2D6 inhibition, estimate hepatotoxicity, estimate human intestinal absorption (HIA), estimate plasma protein binding, assess a broad range of toxicity measures for a set of ligands and remove ligands that are not likely to be drug-like or lead-like.
PHYRE2
Protein HomologY Recognition Engine developed by Lawrence Kelley. PHYRE2 was used for protein structure prediction. It was web-based services for protein structure prediction [9]. PHYRE2 was used to predict the 3-D structure of a protein using the principles and techniques of Homology Modelling, Secondary Structure Prediction and Domain analysis.
SAVS
Stuctural Analysis and Verification Server (SAVS) is a server for analyzing protein structures for validity. SAVS is mainly used for validating the 3D structure of protein whether the protein is stable or not. Ramachandran plot generated by SAVS plays an important role in validating the given 3D structure of protein [19].
3D ligand site searching
3DLigandSite is a web server was used to predict the ligandbinding sites based upon the structure similar to the protein submitted as query. Ligands bound to structures similar to the query were superimposed onto the model. This model used to predict the binding site. 3DLigand Site was used to predict the binding sites using ligands from homologous structure of query protein nsP2 protease [10].
Discovery studio
Discovery studio Package was used for drug design and for protein modeling. It was used for applications such as Catalyst, Modeller, and CHARMm. In the present work, it was used for 3D Pharmacophore screening and for checking the ADMET properties of the ligand molecules [8].
Docking
Docking was used to predict both ligand orientation and binding affinity. In this method the preferred orientation of one molecule with relation to a second, when bound to each other to form a stable complex in three dimensional spaces is predicted [4]. Docking was used for finding the orientation of drugs in particular target. Knowledge of the preferred orientation in turn was used to predict the strength of association/binding affinity between two molecules using scoring functions.
Scoring function
Scoring function was used to predict the binding affinity of one ligand to the receptor molecule. The function of Scoring was used to assess the poses prior to real scoring of the docked complex. Another approach of the scoring function was used to establish a conformational relationship from the large protein databases and then the stability and fitness of the pose was evaluated [6].
Evaluation
Docking score was used to rank the ligands on the basis of their relative binding affinities. Evaluating the results was done by analyzing docked drugs based on their scores [7]. The best docked result was analyzed by the best score of docking that is greatest value of dock score.
GOLD suite
GOLD (Genetic Optimization for Ligand Docking) is a program for calculating the docking models of small molecules in protein binding site and is provided as part of the gold Suite. The GOLD Suite provides all the functionality required to set up individual dockings or virtual screening runs, to post-process docking results and to visualise and manipulate structures in 3D, pre- and post-docking. Raw PDB files can be set-up for use within GOLD. No third party software is needed to prepare the binding site [16] .
An integrated approach to model the nsP2 Protease protein of Chikungunya and to identify the potential nsP2 Chikungunya protease inhibitors, structure based virtual screening has been used in this study. In order to identify novel, safe and effective inhibitor for CHIKV Protease, in-silico screening of competitive inhibitors for nsP2 Chikungunya protease by means of screening compound library for druglikeliness and molecular docking were carried out (Table 1).
Modeling of CHIKV nsP2 protease protein
The three-dimensional structure of a protein nsP2 Chikungunya Protease was predicted by using thePHYRE2. The techniques of Homology Modelling, Secondary Structure Prediction and Domain analysis were used in the process. 320 residues of nsP2 Chikungunya Protease were modelled with 100.0% confidence by the single highest scoring template.
First the nsP2 Protease protein sequence was searched against the Protein Data Bank (PDB). The best template structure was selected based on the sequence homology. Here 2HWK_Chain A, Crystal structure of Venezuelan equine encephalitis2 alpha virus nsP2 Protease domain, shows the highest similarity for the nsP2 Protease Protein of Chikungunya. The template similarity is 41% and remaining protein sequence was structure by principle of domain analysis. 3-Dimensional Structure of CHIKV nsP2 Protease Protein Model is generated using PHYRE2 (Figure 1).
Binding site prediction of CHIKV nsP2 protease
The modelled nsP2 Protease structure was submitted to 3-D Ligand Site Prediction Server. The server found the ligands bound to structures similar to the query and superimposed them onto the model and to predict the binding site. The protein structure library currently used by 3DLigandSite is based on the PDB as of 20 January 2010.
S-adenosyl methionine (SAM) ligand was bound to the structure similar to the nsP2 Chikungunya Protease (Table 3).
| S.No. | Ligand in Structure Similar to the Query Protein | Ligand in PDB Structure Model Similar to the Query Protein |
|---|---|---|
| 1. | SAM (S-adenosyl methionine) |
3dou_A,2pxc_A,2oxt_C,3emb_A,3elw_A, 2wa2_A,3gcz_A |
Table 3: The Ligand present in predicted Binding Site and Ligand present in the PDB model.
The binding sites were predicted based on the binding of SAM to the active. Based on the ligand SAM bound to the structure similar to the Protein CHIKV_nsP2 Protease. The following residues are predicted as best binding sites for the lead compound LEU67, GLU68, ILE85, THR87, PRO88, ARG90, LEU100, LYS103, TYR104, VAL119, ASN132, ILE134, ALA136, ASN137 AND ARG138 (Figure 2).
Ligand screening and preparation
Similarity search and physicochemical property filter: In first step of Virtual screening using Accelerys Discovery Studio Client 2.5, a 2D similarity search for the available inhibitor was done by the PubChem structure search with 85% as threshold value. In this search, totally “11099” analogues were retrieved by using the inhibitor SAM.
Drug likeliness screening: In the reactive functional group screening, 197 library compounds wee filtered from 11099 compounds. The ADMET properties of the compound were analyzed .The compounds that were obeying the drug likeliness property were tabulated (Table 4).
| S.No. | Parameter | Minimum | Maximum |
| 1 | Log P | -2 | 5 |
| 2 | Molecular Weight | 200 | 500 |
| 3 | Hydrogen Bond Acceptors | 0 | 10 |
| 4 | Hydrogen Bond Donors | 0 | 5 |
| 5 | Rotatable Bonds | 0 | 8 |
| 6 | Heavy Atoms | 20 | 70 |
Table 4: Typical range of Physicohemical properties for drug likeliness.
Various steps in the ligand screening and the number of compounds which passed these tests were given in Table 5. Finally 4 ligands having best hits were obtained. From 4 ligands 2 ligands shows good Hydrogen bond interaction and Best Gold Dock Score.
| S.No | Database Name | After 85% of SAM Structure Similarity | After ADMET | Before Drug Likeliness Screening | No. of ligands which passed drug likeliness test | Ligands after HTVS Docking | Best Ligands |
| 1. | Pub Chem Database | 11,000 | 197 | 197 | 90 | 4 | 2 |
Table 5: Ligand Screening Works Flow.
Gold docking result
Gold score for the compound LIGAND_3 was 31.6934 with ADMET_BBB Level -0.712. The binding mode of interaction is shown in Figure 3. Two hydrogen bonds are established during docking of LIGAND_3 with nsP2, Carbonyl Nitrogen atom in the pyrimidine ring of LIGAND_3 forms a hydrogen bond to the carbonyl Nitrogen (N-H…N) of Lys103 and the distance is 2.89Å and Carbonyl Nitrogen atom in the pyrimidine ring of LIGAND_3 forms a hydrogen bond to the Carbonyl Oxygen (N-H…O) of same Residue Lys 103 and the distance is 2.87Å respectively. C10, C15, C18, C20, C23 are involved in hydrophobic contacts. All other residues such as Leu100, Ala136, Asn137 and Arg138 were engaged in non-Ligand hydrophobic interactions. The hydrogen bond interactions are shown using LigPlot (Figure 3).
Gold score for the compound LIGAND_4 was 47.6163 with ADMET_BBB Level -0.481. The binding mode of the compound to active site is shown in Figure 4. One hydrogen bond is established during docking of LIGAND_4 with nsp2. Carbonyl Nitrogen atom in the pyrimidine ring of LIGAND_4 forms a hydrogen bond to the carbonyl Oxygen (N-H…O) of Lys103 the bond distance is 3.00Å. C15, C18, C19, C22, C23, C25, C27, C28, C29, C30, C31, C32 were involved in hydrophobic contacts. All other residues such as Leu100, Tyr104, Val119, Asn132, Ile134, Ala136 and Asn137 were engaged in non- Ligand hydrophobic interactions. The hydrogen bond interactions is shown using LigPlot (Figure 4).
All the ligand interaction with receptor was available in the output file. The best confirmation results were found out using Gold Score, number of Hydrogen bonds formed and Blood Brain Barrier level (Table 6). Among these tow compound LIGAND_4 [N-butyl-9-[3, 4-dipropoxy-5-(propoxymethyl) oxolan-2-yl] purin-6-amine] was found to bind with the target more efficiently than other compound with best gold score(47.6163) and hydrogen bonding interactions with the peripheral site key residue and Blood Brain Barrier level (-0.481). Hence, LIGAND_4 has been emerged as a promising Anti-viral drug candidate with potential symptomatic and disease-modifying effects.
| Name | Structure | Name of the Lead | Gold Score | ADMET_BBB Level |
| LIGAND_3 | 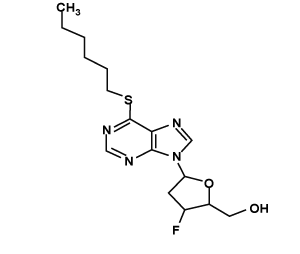 |
[3-fluoro-5-(6-hexylsulfanylpurin-9-yl)oxolan-2-yl]methanol | 31.6934 | -0.712 |
| LIGAND_4 | 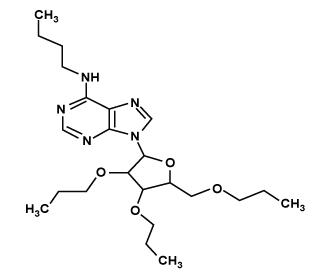 |
N-butyl-9-[3,4-dipropoxy-5-(propoxymethyl)oxolan-2-yl]purin-6-amine | 47.6163 | -0.481 |
Table 6: Results of Docking Analysis for Synthetic compounds.
CHIKV nsP2 protease was responsible for diseases, therefore, identify the drug which inhibits the expression of nsP2 protein prevent the inhibition of interferon during the infection of virus [11]. The nsP2 protease was responsible for cleavage in the nonstructural polyprotein that were crucial for the viral replication cycle. Therefore this nsP2 protease constitutes an attractive target for the development of antiviral compounds [12]. The inhibitor blocks the protease when new viral particle break off from on infected cells. Now days there is no proper inhibitor for inhibit the Chikungunya protease activity.
The study of enzymatic activity of the purified protease nsP2 has ability to cleave peptidyl-7-amino-4-methylcoumarine; this kind of substrate was used to study the activity of a wide range of proteases [13]. Still there was no PDB_ID for the structure of CHIKV nsP2 Protease. Therefore first we retrieve the sequence of nsP2 CHIKV(37997) Protease from UNIPROT_KB and then the protein was Modelled by the query sequence of Venezuelan equine encephalitis virus(VEEV) nsP2 structure [14]. The C-terminal region of alphavirus nsP2 protease could play a similar role to the CHIKV nsp2 protease. The PDB_ID for VEEV virus nsP2 Protease was 2HWK, it shows 40% template similarity for CHIKV nsP2 protease. The Chikungunya protease activity was slightly inhibited by NaCl [15]. But inhibition of CHIKV protease activity by NaCl is not that much effective. Chloroquine [17], Quinine [18], Carbomide, Saquinavir, Indanavir, Ritonavir were the Antiviral drug for CHIKV replication. But these replication compounds were cause some side effects like Dizziness, Nausea, High Blood Sugar, Signs of Allergy, etc. Therefore in our study main was mainly focused to identified the novel inhibitor free from side effect and free from allergy.
Docking work was performed with novel potent inhibitor or lead compound retrieved from PUBCHEM database through screening technique, which fulfill the Drug likeliness and ADMETox property. The Lead compounds were Screened form PUBCHEM Database. Based on PUBCHEM structure similarity search 85% of similar structure of SAM was retrieved. Total number of compound Screened in structure similarity screening was 11099 and the following screened compounds were screened with ADMETox Properties. After this screening 197 Lead compounds were obtained. Then 197 Lead Compound were screened based on Drug Likeliness Property. 4 Best Lead Compound were obtained at last. These 4 ligands were free form side effects because the obtained ligands were screened by ADMETox properties and Physicohemical properties for drug likeliness. Docking work was performed with 4 Lead Compound with the target protein CHIKV nsP2 Protease Protein using GOLD SUITE. Among the 4 compounds, LIGAND_4 has best score (47.6163) compared with other compounds. Hydrogen bonding exists with the residue LYS103 Key residue.
This study describes about the nsP2 Protease that have demonstrated to be a promising drug target for the treatment of Chikungunya. The interaction between Chikungunya nsP2 protease protein against the ligands was studied by using various computational methods. Based on the hydrogen bonds, docking score and Blood Brain Barrier level value the docking result was analyzed. The results were compared within themselves to find out the best ligands which can inhibit the property of the viral protein. Based on this observations LIGAND_4 [N-butyl-9-[3, 4-dipropoxy-5-(propoxymethyl) oxolan-2-yl] purin-6-amine] is found to bind with the target more efficiently than LIGAND_3 compound with best gold score and hydrogen bonding interactions with the peripheral site key residue. Hence, LIGAND_4 has been emerged as a promising Anti-viral drug candidate with potential symptomatic and disease-modifying effects. Further, QSAR studies can be done to identify conformational changes.