Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research Article - (2020)Volume 8, Issue 2
Haemolysis is the lysis of red blood cells (RBCs) and the subsequent release of their contents into surrounding fluid. Several pathogens, including Enterococcus faecalis, Klebsiella pneumoniae, Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa are able to cause haemolysis in vitro and in vivo.
A vast body of scientific literature has demonstrated the ability of certain probiotic bacteria to antagonize grampositive and gram-negative strains by secreting soluble molecules named bacteriocins. Anyway, insufficient data is currently available in relation to haemolytic bacteria.
Ten lactobacilli were selected for this in vitro study. The agar spot assay was employed to quantify any possible inhibition. The diameters of inhibition zones around the spots were measured.
Our results showed that selected probiotics could exert a focused protective effect against pathogenic bacteria responsible for RBCs lysis at various extent. Further investigations will be needed to study the underlying molecules responsible for inhibition.
Haemolytic bacteria; Probiotics; Inhibition activity; Lactobacilli; Pathogens; Bacteriocins
The intestinal microbiota is very well adapted, outstandingly stable and extremely distinct for each individual. In ordinary conditions of stable functioning of the digestive system, neutral and beneficial microorganisms dominate. One hundred trillion (1014) microorganisms are estimated to reside in the human intestine [1,2].
The preservation of a beneficial microbiota necessitates a homeostatic equilibrium within microbial communities, and also between the bacterial entities and the intestinal interface of the host. The resilience of the healthy microbiota offers a protection from dysbiosis-related diseases, such as inflammatory bowel diseases (IBD) or metabolic disorder. By contrast, a resilient dysbiotic microbiota may cause disease [3,4].
Increasing evidence has demonstrated that a permanent alteration in microbiota composition or function (dysbiosis) can modify immune responses, intestinal permeability, metabolism, and digestive motility, thereby creating and perpetuating a proinflammatory state [5].
A huge body of scientific literature has demonstrated the ability of certain probiotic bacteria to antagonize gram-positive and gramnegative strains [6]. The antagonistic activity of a microorganism can be caused by:
•Competitive exclusion
•Immune modulation and stimulation of host defence systems
•Production of organic acids or hydrogen peroxide that lower pH
•Secretion of antimicrobials such as bacteriocins
•Synthesis of signalling molecules that trigger modifications in gene expression [7].
•Short-chain fatty acids (SCFAs) produced during the anaerobic metabolism of carbohydrates have an important role in decreasing luminal pH. The inhibition of microbial growth by organic acids may be due to their ability to pass across the cell membranes, dissociate in the more alkaline environment of the cytoplasm, with consequent acidification [8]. Alternatively, the accumulation of acid anions may cause osmotic stress [9].
•in vitro studies suggest multiple specific activities of different probiotic agents against several pathogens, including Listeria monocytogenes [10], Salmonella typhimurium [11], Escherichia coli [12], and Helicobacter pylori [13] among others. Furthermore, various in vitro and in vivo studies have shown that specific strains of lactobacilli hinder the growth of microbes responsible for bacterial vaginosis, with explicit reference to Gardnerella vaginalis [14].
•During the last few years, a considerable set of scientific evidence has gathered suggesting that specific surface-associated and extracellular components synthesized by probiotic bacteria could be involved in various mechanisms of action. These bacterial components are able to be directly recognized by the host mucosal cells and include bacteriocins, exopolysaccharides, lipoteichoic acids, and surface-associated and extracellular proteins. About 25% to 30% of the bacterial proteins extrinsic their functions in the cell envelope or outside of the cell. Extracellular proteins can be divided into two groups, comprising molecules that are actively transported to the bacterial surroundings through the cytoplasmic membrane, as well as those that are simply shed from the bacterial surface due to the normal turnover of the cell wall. Compared to the other bacterial components, the interactive ability of extracellular proteins or peptides has been less extensively investigated [15].
•Two key features differentiate the majority of bacteriocins from classical antibiotics: The former are ribosomally synthesized, proteinaceous substances with a relatively narrow killing spectrum [16]. Generally, the genes encoding and directing bacteriocin synthesis are organized in operon clusters [17]. They may be located on mobile genetic elements, such as chromosome in association with transposons or plasmids. The bacteriocin family includes a diversity of proteins in terms of size, mode of action, microbial target, release, and immunity mechanisms and can be divided into two main groups, based on production by gram-negative and grampositive bacteria. Secretion of bacteriocins in gram-positive bacteria [18,19] is generally related to the shift from log phase to stationary phase. For example, nisin production begins during mid-log phase and increases to a maximum as the cells enter stationary phase [20]. The regulation of expression is not cell cycle dependent, per se, but rather culture density dependent [21,22]. The most common bacteriocins include lacticin , pediocin , piscicolin , enterocin , reuterin , plantaricin , Enterolysin And nisin [23-30].
•Anyway, the current scientific literature is still deficient of significant data in relation to the possible antagonism of haemolytic microbes by selected biotherapeutic beneficial bacteria. Haemolysis is the lysis of red blood cells (RBCs) and the subsequent release of their contents into surrounding fluid [31]. There are three types of haemolysis, designated alpha, beta and gamma, that could be caused by different pathogenic microorganisms and implying diverse haemolytic reactions observed on blood agar plates [32]. Beta haemolysis (β-haemolysis) consists of a complete lysis of red cells in the media around and under the colonies. Streptolysin, an exotoxin, is the enzyme synthesized by the bacteria causing the complete lysis of RBCs [33]. On the other side, alpha haemolysis is also denominated as incomplete haemolysis or partial haemolysis. It is caused by the hydrogen peroxide secreted by specific bacteria, with consequent haemoglobin oxidization and production of the green oxidized derivative methaemoglobin [34].
•Haemolysis occurs normally in a small percentage of red blood cells as a means of removing aged cells from the bloodstream and freeing heme for iron recycling [35,36]. It also can be induced by exercise [37]. The ability of bacterial colonies to induce haemolysis when cultured on blood agar is used to classify certain microorganisms. A substance that causes haemolysis is a hemolysin.
•Our study focused on the inhibition assessment of Enterococcus faecalis ATCC 19433 (γ-haemolysis), Klebsiella pneumoniae ATCC 700603, Escherichia coli ATCC 8739 (α-haemolysis), Staphylococcus aureus ATCC 25923 (β-haemolysis), and Pseudomonas aeruginosa ATCC 9027 by ten selected probiotic bacteria.
Bacterial strains and culture conditions. In the present study, ten lactobacilli were selected, namely L. acidophilus LA02 (DSM 21717), L. plantarum LP14 (DSM 33401), L. plantarum LP01 (LMG P-21021), L. reuteri LRE02 (DSM 23878), L. salivarius LS01 (DSM 22775), L. acidophilus LA06 (DSM 23033), L. fermentum LF26 (DSM 33402), L. casei LC04 (DSM 33400), L. paracasei LPC09 (DSM 24243), and L. rhamnosus LR04 (DSM 16605). They were all obtained from the internal bacteria collection of Probiotical SpA, Novara, Italy. Prior to their use according to the protocol, the bacteria were subcultured twice in De Man, Rogosa and Sharpe broth (MRS) (Becton Dickinson, Milan, Italy) at 37°C for 18 hours, using an anaerobic atmosphere generation system (GasPak) (Becton Dickinson, Milan, Italy).
The haemolytic bacteria Enterococcus faecalis ATCC 19433, Klebsiella pneumoniae ATCC 700603, Escherichia coli ATCC 8739, Staphylococcus aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 9027 came from the American Type Culture Collection (ATCC, Rockville, Maryland, USA). They were sub-cultured twice in Brain Heart Infusion (BHI) broth (BiolifeItaliana Srl, Milan, Italy) at 37°C for 18 h. The liquid cultures of pathogens were brought at the Optical Density (OD) equal to n. 2 of McFarland scale using BHI broth as diluent. All the strains were sub-cultured at least three times before being used in the inhibitory experiments.
Inhibition assay: The agar spot assay was employed to quantify any possible inhibition according to the protocol described by Santini, et al. [38]. This method consisted of the following steps: MRS agar was preparedand 5 μl of the probiotic bacteria fresh cultures with an optical density (OD) at 600 nm around 1 were spotted on the surface. The plates were then incubated anaerobically for 5 h at 37°C in order to allow the growth of the spots.
Next, the pathogenic bacteria suspensions were inoculated at 4% in 5 ml of BHI soft agar and after homogenization they were poured over the plates previously seeded with probiotics. When the top agar was solid, the plates were inverted and incubated aerobically for 24 h and 48 h at 37°C. At the end of incubation, the inhibition zones were recorded, when applicable.
Results were expressed as the mean diameter of inhibition halos (mm) ± Standard Deviation (SD). The experiments were repeated three times for each combination Lactobacillus/haemolytic bacterium.The cut-off for a result to be regarded as significant was 1 mm.
Lactobacillus plantarum LP14 showed the strongest direct inhibition activity on the majority of the haemolytic targets tested, with inhibition areas measuring on average 10 mm. The only exception was represented by Enterococcus faecalis ATCC 19433, against which L. rhamnosus LR04, L. fermentum LF26 and L.casei LC04 demonstrated the most significant evidence (Figure 1).
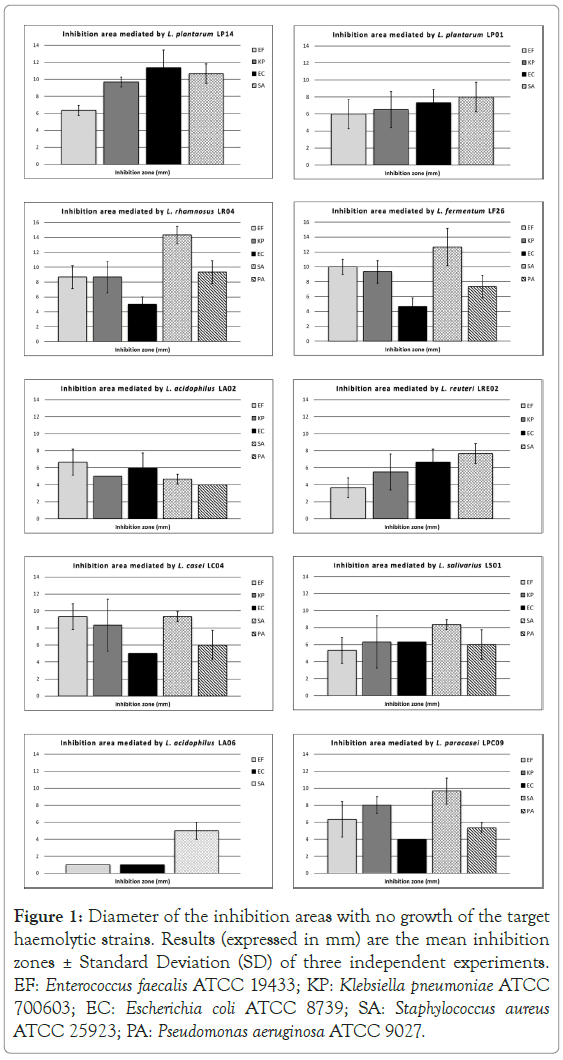
Figure 1. Diameter of the inhibition areas with no growth of the target haemolytic strains. Results (expressed in mm) are the mean inhibition zones ± Standard Deviation (SD) of three independent experiments. EF: Enterococcus faecalis ATCC 19433; KP: Klebsiella pneumoniae ATCC 700603; EC: Escherichia coli ATCC 8739; SA: Staphylococcus aureus ATCC 25923; PA: Pseudomonas aeruginosa ATCC 9027.
L. salivarius LS01 and L. plantarum LP01 were effective as well, with average inhibition diameters between 6 and 9 mm.
The individual strongest antagonism towards Staphylococcus aureus ATCC 25923 was reported when using L. rhamnosus LR04 (14.33 ± 1.15 mm of mean inhibition halo) and L. fermentum LF26 (12.67 ± 2.52 mm of mean inhibition area) (Figure 1).
On the other side, albeit L. reuteri LRE02 is able to synthesize the molecule, the inhibition results against haemolytic bacteria are slightly lower if compared with the majority of the other lactobacilli, even if well above the 1 mm threshold.
L. acidophilus LA06 demonstrated the lowest activity, even if this lactobacillus was tested only against three pathogens.
The antimicrobial activity of probiotic microorganisms has a very wide area of application including protective effects in case of urinary tract infections (UTIs), adjuvant therapy to antibiotic treatment and gastrointestinal tract associated diarrhea, prevention of nosocomial infections, eradication of H. pylori and dental biofilm formation, as well as in the agro‐food industry for manufacturing fermented products.
The process of determining antimicrobial features of probiotics is multifaceted and includes in vitro assays, in vivo models or substitute models, clinical studies, metagenomic analyses and mathematical modelling.
There is an explicit need for more elaborate assays able to better signify the complex interactions between the probiotics and the host microbiome to comprehend the consequences of the in-situ production of antimicrobials.
The potential clinical application of bacteriocins produced by Lactic Acid Bacteria (LAB) has been recently the subject of investigations by many scientists. Bacteriocins may be regarded in a sense as antibiotics, even though they differ from conventional molecules in numerous aspects.
Our in vitro study demonstrated that selected probiotics could exert a focused protection effect against pathogenic bacteria responsible for RBCs lysis at various extent.
Further investigations from our group highlighted that the chromosomal sequences of eight out of ten lactobacilli tested evidence the presence of genes coding for bacteriocins. More specifically, L. salivarius LS01 contains two genes coding for Enterolysin A and Salivaricin P chain b. An article by Barrett, et al. suggested that this two-component bacteriocin may be a common feature of intestinal L. salivarius strains [39]. L. plantarum LP14 incorporates a gene coding for Plantaricin J, L. reuteri LRE02 has a putative gene for the synthesis of Enterolysin A, L. acidophilus LA02 encompasses at least three interesting sites (Acidocin J, Enterolysin A and Helveticin J), L. plantarum LP01 has a putative site for the synthesis of Plantaricin E, L. rhamnosus LR04 genome contains a site coding for Carnocin-CP52 [40], L. casei LC04 includes three interesting sites (Carnocin-CP52, Enterocin Xβ, Listeriocin 743A), and L. paracasei LPC09 contains two genes encoding for LSEI_2386 and Thermophilin A. On the other side, L. fermentum LF26 seems to contain no putative genes coding for bacteriocins, even if it was one of the most effective strains in term of haemolytic pathogens inhibition (unpublished results). Additional analysis will be needed with reference to this probiotic strain.
L. acidophilus LA06 recorded the lowest antagonistic activity, even if it was tested only against three pathogens. This evidence most probably has to be attributed to its homo fermentative metabolism and the sole secretion of organic acids in the surrounding microenvironment.
With this regard, it is interesting to point out that the ability to secrete a significant amount of organic acids is a desirable and useful feature in relation to a biotherapeutic activity. A recent study by Sorbara,et al. demonstrated that an antibiotic-naive microbiota markedly hinders the growth of antibiotic-resistant clinical isolates of Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis by acidifying the proximal colon and triggering short chain fatty acid (SCFA)-mediated intracellular acidification. In this way, the inhibition of Enterobacteriaceae is completely dependent on an acidified pH. Coupled with the production of high levels of SCFA, this acidified environment generates intracellular acidification of Enterobacteriaceae to an extent able to prevent replication [41].
Besides the mere acidification, the ability of a biotherapeutic bacterium to synthesize and secrete one or more bacteriocins may reinforce its overall effectiveness and confer a more focused inhibition targeting.
The whole body of experimental evidence is absolutely consistent with the results ranking emerged during the present analysis.
Further investigations will be needed to quantify the secretion of each antimicrobial molecule discovered at the chromosome level as well as to hypothesize and define some potential applications of these probiotics aimed at offering a targeted protection against haemolytic pathogens in different areas.
Nina Vinot contributed with the English proofreading of the manuscript.
The study has been entirely funded by Probiotical SpA.
Our research did not include any human subjects and animal experiments since it was an in vitro investigation.
Marco Pane, Angela Amoruso, Francesca Deidda, Teresa Graziano, Annachiara De Prisco and Luca Mogna are employees of Probiotical Research Srl (Novara, Italy).
Mario Del Piano is the Head of Clinical Research of Probiotical SpA (Novara, Italy).
Citation: Deidda F, Graziano T, Amoruso A, De Prisco A, Marco P, De Prisco A (2020) How Probiotics may Kill Harmful Bacteria: The in vitro Activity against Some Haemolytic Strains. 8:216.
Received: 18-Mar-2020 Accepted: 10-Apr-2020 Published: 17-Apr-2020 , DOI: 10.35248/2329-8901.20.8.216
Copyright: �?�© 2020 Deidda F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : The study has been entirely funded by Probiotical SpA