Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2024)Volume 17, Issue 3
This study endeavors to evaluate the potential pathogenic or benign consequences of the Liver Kinase B1 (LKB1) protein mutation, Mitogen Activated Protein Kinase p38alpha (D176A), which is categorized as a Variant of Unknown Significance (VUS). LKB1 serves as a widely recognized tumor suppressor protein with multifaceted involvement in cellular processes such as metabolism regulation and apoptosis induction. It forms a heterotrimeric complex that becomes activated through interaction with STe20-Related Adaptor (STRAD) and Mouse protein 25 (MO25). Peutz-Jeghers syndrome, an autosomal dominant hereditary disorder, is associated with mutations in the LKB1 gene. Existing evidence suggests a high likelihood of deleterious effects attributed to the amino acid substitution D176A, as inferred from the outcomes of nine out of ten predictive models. The conservation analysis conducted through the Conserved Domain Database (CDD) and aminode reveals notable preservation of the aspartate residue at position 176 across many diverse species. Despite the absence of statistically significant differences in the Root-Mean-Square Deviation (RMSD) data between the native and mutant LKB1 structures, assessments utilizing Yet another Scientific Artificial Reality Application (YASARA) indicate a diminished binding affinity of the variant LKB1 to the STRAD protein compared to its native counterpart. Comparing the native and variant LKB1 tertiary structures showed misalignments upon superimposition.
RMSD data of various specific amino acid residues exhibited disparities of more than 2 angstroms. Additionally, discernible variances in the positioning of cavity pockets are observed between the native and variant LKB1 proteins. Collectively, the analytical findings suggest that the D176A mutation in the LKB1 protein is likely to entail detrimental consequences.
Tumor suppressor protein; Clinvar; STK11; Peutz-jegers; LKB1
The protein denoted as Serine/Threonine Kinase 11 (STK11) or LKB1 assumes a pivotal role in diverse cellular phenomena encompassing energy sensing pathways, cellular metabolism, cell polarity, apoptosis and Deoxyribonucleic Acid (DNA) damage response mechanisms [1]. Particularly noteworthy is its interplay with the Tumor-suppressor Protein (p53), which serves as a significant mediator in the regulation of cellular apoptosis [1]. Activation of LKB1 is contingent upon conformational alterations facilitated by its interaction with two auxiliary proteins, namely STRAD and MO25 [1,2]. The catalytically active site of LKB1 harbors an aspartate residue at position 176, which functions as a proton acceptor during phosphorylation events [3]. Notably, the Adenosine Triphosphate (ATP) binding site is situated within the amino acid residues spanning 55-78 amino acids [4]. Germline mutations within the LKB1 gene precipitate the clinical manifestation of Peutz-Jeghers Syndrome, an autosomal dominant disorder characterized by the development of hamartomatous polyps within the gastrointestinal tract. Furthermore, accumulating evidence implicates somatic mutations within the STK11 gene in the etiology of diverse malignancies, encompassing gastrointestinal cancers, pancreatic cancer, cervical cancer, melanoma and non-small cell lung cancer, thereby emphasizing its multifaceted role in tumorigenesis [5]. LKB1's role as a tumor suppressor gene is underscored by its functional impairment, which elicits dysregulated cellular proliferation, heightened macromolecular synthesis and augmented aerobic glycolysis-phenotypic hallmarks of malignant transformation [1]. An illuminating investigation conducted on a Chinese familial cohort afflicted with Peutz-Jeghers Syndrome elucidated pertinent clinical and molecular insights [6]. The proband, together with her maternal lineage, exhibited mucocutaneous pigmentation, subsequently confirmed by endoscopic evaluation revealing the presence of gastrointestinal polyps [6]. Molecular analysis identified a novel mutation within exon 4 of the STK11 gene, precipitating a heterozygous alteration at amino acid residue 174 from histidine to proline [6]. Employing Swiss-Modeling techniques, the study delineated the impact of this mutation on hydrogen bonding patterns involving critical amino acids, including residues 174, 193, 194, 176 and 237, underscoring its pathogenic significance [6].
In this study, a human gene, specifically LKB1/STK11 D176A variant, was selected for investigation utilizing data from ClinVar, a repository provided by the National Institutes of Health (NIH) [7]. The LKB1 protein structure was obtained from multiple sources including Uniprot, Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PBD) and Iterative Threading ASSEmbly Refinement (I-TASSER), with emphasis placed on the I-TASSER dataset for subsequent analyses [8,9]. Employing YASARA software, a series of procedural steps were undertaken including structure refinement through cleaning and energy minimization [10]. These steps comprised executing the em_runclean macro, generating pdb/yob files for the Variant of Unknown Significance (VUS), hydrogen addition, optimization of hydrogen bonding and creation of replicate pdb/yob files followed by energy minimization. The native and D176A replicate proteins were run via computer simulations. Additionally, resources such as aminode and conserved domain databases were utilized.
Further investigation into the pathogenic potential of the D176A variant was conducted utilizing nine predictive programs including predict Single Nucleotide Polymorphism (SNP), Multivariate Analysis of Protein Polymorphism (MAPP), Predictor of human Deleterious Single Nucleotide Polymorphisms (PhD SNP), Polyphen-1, Polyphen-2, Sorting Intolerant From Tolerant (SiFT), Single Nucleotide Polymorphism Annotation Platform (SNAP), Evolutionary model of Variant Effect (EVE), SNP Effect Predictor (SNAP2) and Protein Analysis Through Evolutionary Relationships (PANTHER). Subsequently, an in-depth analysis of simulation data was performed encompassing statistical comparisons, specifically Root Mean Square Deviation (RMSD) analysis between native and variant LKB1 proteins, along with statistical significance assessment through p-values. Furthermore, RMSD comparisons were conducted with a focus on specific amino acid residue sites, alongside correlational analysis depicted through heat mapping techniques.
The YASARA software facilitated various analytical procedures including superimposition of native and variant LKB1 proteins, analysis of binding affinities between native/variant LKB1 and associated proteins STRAD, MO25 and Atrial Natriuretic Peptide (ANP), as well as graphical representation through figures. Additionally, molecular cavity surface comparisons were conducted between native and variant LKB1 proteins to elucidate potential structural alterations [5-10].
Nine out of ten predictive programs suggest that the D176A variant of unknown significance could have a deleterious impact (Table 1). Additionally, analyses from the Conserved Domain Database (CDD) and aminode reveal that the aspartate at the 176 amino acid position is highly conserved across 33 different species, including Homo sapiens. YASARA's superimposition of native and variant LKB1 proteins highlights significant misalignments across various sections of their structures, with notable deviations emphasized in yellow based on RMSD data (Figure 1). Highlighted in yellow shows residues with more than 2 Å differences between the native and variant LKB1 protein. Highlighted in orange is the Variant of Unknown Significance (VUS) D176A. Furthermore, the variant LKB1 exhibits a reduced binding affinity to its partner STRAD, interacting with only 19 amino acid residues-a 77% decrease compared to the native LKB1 (Table 2). Moreover, differences in molecular cavity patterns around the 176 amino acid residues further distinguish the native and variant LKB1 proteins, orange cavity pockets for blue variant LKB1 protein and yellow cavity pockets for gray native LKB1 protein. Highlight in pink shows D176 for the Native LKB1 and D176A for the variant LKB1 protein (Figure 2).
| Predictive program | Results |
|---|---|
| Aminode | Region of Variant of Unknown Significance (VUS) 163-186 highly conserved in many species, substitution score of 0 |
| Single Nucleotide Polymorphism (SNP) | Deleterious 79% confidence |
| Multivariate Analysis of Protein Polymorphism (MAPP) | Deleterious 91% confidence |
| Predictor of human Deleterious Single Nucleotide Polymorphisms (PhD SNP) | Neutral 45% confidence |
| PolyPhen-1 | Deleterious 74% confidence |
| Polyphen-2 | Deleterious 81% confidence |
| Sorting Intolerant From Tolerant (SiFT) | Deleterious 79% confidence |
| Single Nucleotide Polymorphism Annotation Platform (SNAP) | Deleterious 85% confidence |
| Evolutionary model of Variant Effect (EVE) | Pathogenic, EVE score of 0.749 |
| SNP Effect Predictor (SNAP2) | Effect-score of 89-91% confidence |
| Protein Analysis Through Evolutionary Relationships (PANTHER) | Probably damaging, Pdel 0.89 |
Table 1: Predictive program results in which 9/10 indicate likely pathogenic effects with a mitogen activated protein kinase p38alpha (D176A) mutation in the Liver Kinase B1 (LKB1) protein.

Figure 1: Yet Another Scientific Artificial Reality Application (YASARA) superimposed variant Liver Kinase B1 (LKB1) (gray) on native LKB1
protein (blue).  Residues with more than 2 Å difference between native and variant proteins.
Residues with more than 2 Å difference between native and variant proteins.
| Binding affinity | Binding energy (Kcal/mol) | Dissociation constant (mM) | % of residues |
|---|---|---|---|
| Native LKB1 with STRAD | 0.892 | 221.9 | 83 |
| Variant LKB1 with STRAD | 0.274 | 629.73 | 19 |
Table 2: Yet Another Scientific Artificial Reality Application (YASARA) binding affinity of native/variant Liver Kinase B1 (LKB1) with STe20-Related Adaptor (STRAD) protein.
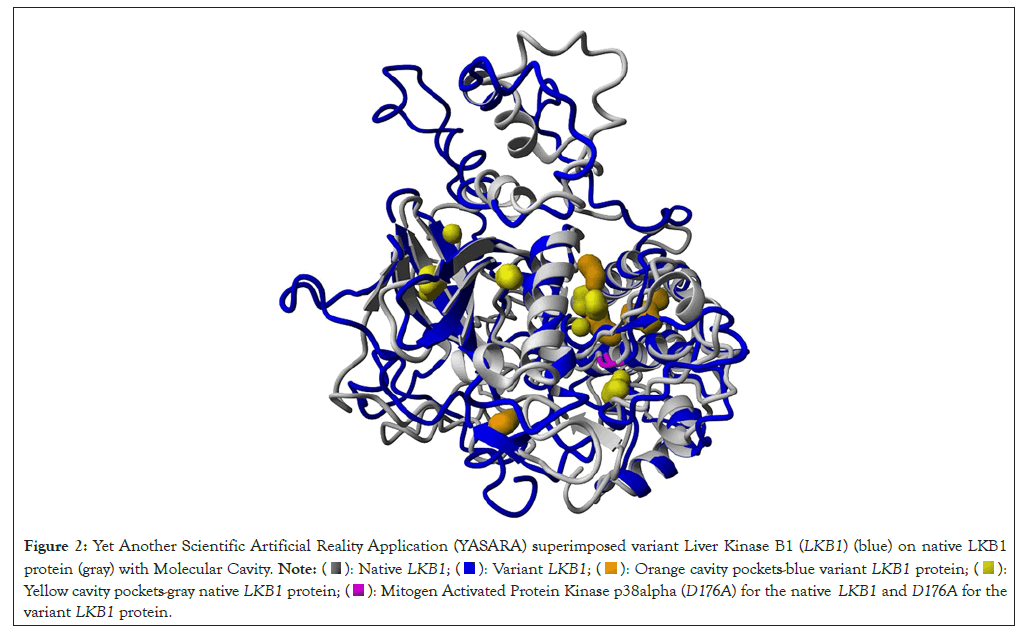
Figure 2: Yet Another Scientific Artificial Reality Application (YASARA) superimposed variant Liver Kinase B1 (LKB1) (blue) on native LKB1
protein (gray) with Molecular Cavity.  Orange cavity pockets-blue variant LKB1 protein;
Orange cavity pockets-blue variant LKB1 protein;  Yellow cavity pockets-gray native LKB1 protein;
Yellow cavity pockets-gray native LKB1 protein;  Mitogen Activated Protein Kinase p38alpha (D176A) for the native LKB1 and D176A for the
variant LKB1 protein.
Mitogen Activated Protein Kinase p38alpha (D176A) for the native LKB1 and D176A for the
variant LKB1 protein.
The conservation of the amino acid residue aspartate 176, D176; across diverse species underscores its critical role within the catalytic domain of the LKB1 enzyme. Positioned as a proton acceptor, D176 plays a pivotal role in maintaining the structural integrity and functional activity of LKB1. The substitution of this polar aspartate with a non-polar alanine residue, as seen in the D176A variant, is likely to induce structural alterations that could affect protein conformation, ligand binding capacity and overall functional activity. The reduced affinity observed between the variant LKB1 carrying the D176A mutation and its partner STRAD further supports the potential functional consequences of this mutation.
However, it is important to acknowledge the limitations of this study. Computational simulations and predictive analyses provide valuable insights into potential structural and functional changes induced by the D176A variant. Yet, these findings necessitate experimental validation using biochemical assays and cellular models to confirm the actual impact on LKB1 activity and downstream signaling pathways. Additionally, the structural modeling presented here relies on computational predictions, which may not fully capture the complexity of protein dynamics observed in vivo.
Future research should aim to elucidate the precise molecular mechanisms through which the D176A variant alters LKB1 function. Experimental studies could explore the effects of this mutation on LKB1 kinase activity, substrate specificity and its role in regulating key signaling pathways such as Adenosine Monophosphate-Activated Protein Kinase (AMPK) and Mammalian Target of Rapamycin (mTOR). Moreover, investigating the broader implications of LKB1 dysfunction in disease contexts, particularly in cancer and metabolic disorders, could uncover novel therapeutic strategies.
Enhanced screening modalities, including genetic profiling in larger cohorts and diverse populations, would provide a clearer understanding of the prevalence and clinical significance of the D176A variant. Integration of multi-omics approaches, such as genomics and proteomics, could offer comprehensive insights into the molecular pathways influenced by LKB1 and its variants.
In summary, while this study highlights the potential pathogenicity of the D176A variant in LKB1, further experimental validation and exploration are essential. A deeper understanding of LKB1's regulatory roles and its influence on cellular signaling cascades holds potential for informing targeted therapies and personalized medicine approaches in the management of diseases associated with LKB1 dysfunction.
In conclusion, our study underscores the critical role of the conserved aspartate residue at position 176 in the LKB1 protein. The Mitogen Activated Protein Kinase p38alpha (D176A) variant, identified through computational and structural analyses, demonstrates potential implications for protein function, including altered binding affinity and structural integrity. These findings highlight the importance of further experimental validation to elucidate the precise impact of this variant on LKB1-associated pathways and its potential relevance in disease contexts. Continued research in this area promises to enhance our understanding of LKB1 biology and may ultimately inform targeted therapeutic strategies for diseases influenced by LKB1 dysfunction.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Cid J (2024). D176A Variant in the LKB1/STK11 Tumor Suppressor Protein: Potential Pathogenic Implications. J Proteomics Bioinform. 17:670.
Received: 02-Aug-2024, Manuscript No. JPB-24-33140; Editor assigned: 05-Aug-2024, Pre QC No. JPB-24-33140 (PQ); Reviewed: 19-Aug-2024, QC No. JPB-24-33140; Revised: 26-Aug-2024, Manuscript No. JPB-24-33140 (R); Published: 02-Sep-2024 , DOI: 10.35248/0974-276X.24.17.670
Copyright: © 2024 Cid J. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.