Research Article - (2016) Volume 2, Issue 2
Identification and Characterization of a New Enterobacter Onion Bulb Decay Caused by Lelliottia amnigena in China
*Corresponding Author: Jianuan Zhou, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Department of Plant Pathology, South China Agricultural University, Guangzhou, 510642, PR China, Tel: +86 20 3863 2491 Email:
Abstract
Objectives: Enterobacter is a genus with numerous species associated with clinical relevance, plants, foods and environmental sources. However, the taxonomy of Enterobacter is complicated and confusing. In order to identify the taxonomy of the causal agent isolated from decay onion bulbs in China and provide an example to identify plant pathogens, we used biochemical technologies in combination with molecular biology to confirm the status of the pathogen.
Methods: In this study, biochemical test, colonial and microscopic morphology analysis, 16S rRNA gene sequencing and multilocus sequences analysis (MLSA) based on partial sequencing of rpoB, atpD, gyrB and infB were performed on the isolates obtained from the decayed onion bulbs. According to the biochemical characteristics and genetic relationship, we compared the pathogen with related strains obtained from BLASTn alignment. Finally, pathogenicity test was performed on healthy onion bulbs to verify the Koch’s postulates.
Results: Biochemical test, colonial and electron microscopic morphology indicated that the pathogen is gramnegative, belongs to the genus Lelliottia and in combination with phylogenetic analysis, it is most similar to L. amnigena.
Conclusion: To our knowledge, this is the first report of L. amnigena naturally causing soft rot disease on harvested onion.
Keywords: Onion bulb; Decay; Pathogen; Enterobacter; Lelliottia; MLSA; Identification
Introduction
Enterobacteriacae is a large family of Gram-negative bacteria including many familiar pathogens such as Escherichia, Enterobacter, Erwinia, Dickeya, Citrobacter, Klebsiella, Pantoea, Proteus, Salmonella, Serratia and Shigella. Members of the Enterobacteriaceae can be trivially referred to as enterobacteria or "enteric bacteria" and the type species is Escherichia coli, causing intestine disease in animals.
Since created in 1960, Enterobacter has become one of the largest genera within Enterobacteriaceae, usually lives in almost all habitats, including in normal intestinal flora, in stool of animals, in plants, in water, in insects and in foods. However, some Enterobacters were founded to be phytopathogens, e.g. E. cloacae and E. mori both caused bacterial wilt of mulberry in China [1,2]. E. cloacae also caused some post-harvest plant diseases such as onion bulb rot in the Columbia basin of Washington state [3], ginger rhizome rot in Brazil [4] and papaya necrosis [5]. L. nimipressuralis (formerly named as E. nimipressuralis) caused elm wet wood [6], E. cancerogenus resulted in poplar canker and E. dissolvens infected corn and resulted in maceration rot [5].
Enterobacter has a long and complicated taxonomic history and currently includes 19 species [7]. However, with the development of taxonomy and identification methods, many species were reclassified from this genus, e.g. E. agglomerans was transferred from Enterobacter to the genus Pantoea [8]. E. intermedius was reassigned to Kluyvera intermedia [9]. E. sakazakii was transferred to the genus Cronobacter [10] and more recently, E. nimipressuralis and E. amnigenus were proposed to reclassify into a novel genus Lelliottia as L. nimipressuralis and L. amnigena, respectively, E. gergoviae and E. pyrinus into Pluralibacter as P. gergoviae and P. pyrinus, respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia as K. cowanii, K. radicincitans, K. oryzae and K. arachidis, respectively and E. turicensis, E. helveticus and E. pulveris into Cronobacter as C. zurichensis, C. helveticus and C. pulveris, respectively [7].
Currently, 16S rRNA gene sequence is found insufficient to assign the taxonomy of new species owing to its polyphyletic nature not only in a same family, but also in a same genus. Even the multilocus sequences analysis (MLSA) based on common house-keeping gene sequences such as rpoA, rpoB, recN and thdF resulted in confusing conclusions on the taxonomy of the enterobacteria [11]. Thereafter, an effective combination of methods should be proposed to assign a new species in taxonomy based on traditional observation technologies such as phenotypic characteristics and modern molecular biological means such as DNA-DNA hybrids, 16S rRNA gene sequence analysis, MSLA and conserved gene cluster analysis.
In our study, we identified a pathogen isolated from decayed onion bulbs as L. amnigena according to the cultured colonial morphology, microscopic morphology, biochemical characteristics, combining with the analysis of 16S rRNA gene sequence and the MSLA based on the partial sequences of rpoB, atpD, gyrB and infB, suggesting to provide a reference to assign the taxonomy of the pathogen identification.
Materials and methods
Pathogen isolation
On Nov 22th, 2014, several rot samples of onion bulbs were collected from a local market in Guangzhou city, Guangdong province, China. Scales from four diseased bulbs were surface-sterilized in 70% ethanol for 30 s and rinsed 3 times in sterilized water and cut into small pieces (1 to 5 mm in length). Tissues were then macerated for 5 min in sterilized water and the lixivium was streaked onto Luria- Bertani (LB) medium plates and incubated at 28°C for 24 hours [12]. Cultures were maintained frozen at -80°C in Luria-Bertani (LB) supplemented with 20% (v/v) glycerol.
DNA extraction and PCR amplification
Bacterial genomic DNA was extracted using the MasterPure™ DNA Purification Kit following the manufacturer’s protocol (Epicentre Co., USA) and stored at -20°C. Primers used for amplification of the 16S rRNA gene were 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGCTACCTTGTTACGACTT-3’) with conditions determined by Coenye et al. [13]. Partial sequences of the RNA polymerase β subunit (rpoB) was amplified using primers rpoB CM7-F (5’-AACCAGTTCCGCGTTGGCCTG-3’) and rpoB CM31b-R (5’- CCTGAACAACACGCTCGGA-3’), while DNA gyrase subunit B (gyrB) using gyrB 01-F (5’- TAARTTYGAYGAYAACTCYTAYAAAGT-3’) and gyrB 02-R (5’- CMCCYTCCACCARGTAMAGTT-3’), initiation translation factor 2 (infB) using infB 01-F (5’-ATYATGGGHCAYGTHGAYCA-3’) and infB 02-R (5’-ACKGAGTARTAACGCAGATCCA-3’) and ATP synthase β subunit (atpD) using atpD 01-F (5’- RTAATYGGMGCSGTRGTNGAYGT-3’) and atpD 02-R (5’- TCATCCGCMGGWACRTAWAYNGCCTG-3’) with conditions previously described [14]. Amplicons were purified with Omega’s EZNA. TM Cycle Pure Kit following the manufacturer’s protocol (Omega, USA) and sequenced by Invitrogen Company in Guangzhou, China. The sizes of the resultant amplicons were: rpoB =637 bp, gyrB =742 bp, infB =615 bp and atpD =642 bp.
Multilocus sequences analysis (MLSA) and phylogenetic analysis
Sequences analyses were performed using DNASTAR Lasergene SeqMan program. Sequence similarities of 16S rRNA gene were determined using EzTaxon-eserver (http://www.ezbiocloud.net/eztaxon) [15] and BLASTn program. Sequences of rpoB, gyrB, infB and atpD genes of related strains were obtained from GenBank database. Sequences were aligned with ClustalW and phylogenetic trees were reconstructed using the neighbor-joining method with maximum composite likelihood model. Bootstrap analysis was performed based on 1000 replicates. The MEGA5 package was used for all phylogenetic analyses [16]. The GenBank accession numbers of the 16S rRNA, rpoB, gyrB, infB and atpD gene sequences of strain ZJN are KP642511, KP642510, KP642508, KP642509 and KP642507, respectively.
Colonial and microscopic morphology
Colonial morphology was observed on LB agar plate with bacteria grown for 8 h, the bacterial cells were suspended in sterile distilled water and stained with phosphotungstic acid [3% (v/V), pH 7.0] for 2 min, air-dried and observed by using transmission electron microscope (Hitachi H7650).
Biochemical test
Phenotypic characterization was carried out according to the minimal standards proposed by Brady et al. [7]. Biochemical features of the isolate were studied using standardized procedures [17] of the following tests: Gram staining, oxidase, catalase, cell motility, oxidation and fermentation test, gas and acid production from glucose, indole, methyl red, Voges-Proskauer reaction, utilization of citrate, propanedioic acid, hydrogen sulfide, arginine dihydrolase, lysine and ornithine decarboxylases, urease, O-nitrophenyl-β-Dgalactopyranoside, nitrate reduction and gelatine hydrolysis. The Biolog system (GEN III v2.7.1.40.I5G) was used to determine the carbohydrate fermentation profile and readings were made after 22 h of incubation at 33°C.
Pathogenicity test on bulbs
For the pathogenicity test, mature bulbs were chosen as a randomized complete block design with a factorial combination of the red cultivars (Redwing representing common red storage cultivars grown in the Shandong province of China). Three inoculation treatments [strain ZJN, Escherichia coli DH5α and Phosphate-buffered saline (PBS) buffer] in three inoculation locations were performed. Strains ZJN and E. coli DH5α were grown overnight in 15 ml of LB medium at 30°C and 37°C respectively with 200 rpm oscillation. Cells were harvested by centrifugation (3000 rpm for 5 min), washed with 0.5-fold volume of PBS buffer and resuspended in PBS buffer to an optical density at 600 nm of 0.3-0.5 (approximately 1×108 CFU/ml) and 0.2 ml inoculum or PBS buffer was injected into each bulb [18,19]. The inoculated bulbs were kept at 30°C for approximately three weeks. Three replicates of three onion bulbs were used for each treatment.
Results
Symptoms of the onion bulb decay disease
In this study, we collected decay onion bulbs from a local market and found that the naturally infected bulbs showed intact onion on peel, but decayed internal tissues and odorous smells dispersed when cutting open. After disease development, the whole bulbs became soft, watery and decayed (Figure 1A).
Figure 1: Symptoms of natural infection and microscopic morphology of the isolate. (A) Infected bulbs showed intact onion on peel, but decayed internal tissues and odorous smells dispersed when cutting open. (B) Transmission electron micrograph of isolate ZJN showed that cells are straight rod, with peritrichous flagella and fimbriae and wrinkle cell surface. Bar=200 nm.
Morphological characteristics of the casual agents
Casual agents from the interfaces of health and disease were isolated and grown on LB agar plates. Colonies of the cultured bacteria seemed approximate and eight representative isolates were selected for further characterization. Cultural results showed that the colonies were unpigmented, convex, round and smooth with entire margins. Electron micrograph showed that the bacterial cells are straight rod, 0.7-1.0 μm × 0.88-2.68 μm in size, with peritrichous flagella and fimbriae and wrinkle cell surfaces (Figure 1B).
Phylogenetic analysis on the strain
Then 16S rRNA gene sequence analysis and MLSA were conducted for rapid classification of the strains. Results showed that all sequences of the eight isolates were identical in each gene. Analysis on the 16S rRNA gene sequence revealed strain ZJN belonged to the Enterobacteriaceae, showing the similarities with Enterobacter aerogenes (99.43%), Lelliottia amnigena (99.35%), Kluyvera cryocrescens (99.27%), Enterobacter soli (99.07%), Leclercia adecarboxylata (98.85%), L. nimipressuralis (98.63%) and Buttiauxella izardii (97.85%) (Table 1).
| Gene sequence similarity (%) with strain ZJN | ||||||
|---|---|---|---|---|---|---|
| Species | 16S rRNA | rpoB | atpD | gyrB | infB | Concatenated rpoB, atpD, gyrB and infB |
| Enterobacter aerogenes | 99.43 | 95.29 | 93.77 | 88.68 | 87.97 | 91.42 |
| Enterobacter cloacaesubsp. cloacae | 97.84 | 95.92 | 94.08 | 86.39 | 88.29 | 91.17 |
| Enterobacter cancerogenus | 98.99 | 95.76 | 94.7 | 88.41 | 89.27 | 92.04 |
| Kluyvera cryocrescens | 99.27 | 95.13 | 93.61 | 87.73 | 90.08 | 91.64 |
| Enterobacter soli | 99.07 | 95.45 | 93.77 | 86.39 | 90.41 | 91.51 |
| Lelliottia amnigena | 99.35 | 92.94 | 94.55 | 88.27 | 92.85 | 92.15 |
| Lelliottia nimipressuralis | 98.63 | 93.25 | 94.55 | 89.08 | 92.85 | 92.43 |
| Leclerciaadecarboxylata | 98.85 | 96.23 | 95.02 | 87.60 | 92.52 | 92.84 |
Table 1: 16S rRNA, rpoB, atpD, gyrB and infB gene sequence similarities between strain ZJN and type strains of phylogenetically related species.
Phylogenetic analysis based on 16S rRNA gene sequence with neighbor-joining approach indicated that the isolate ZJN was clustered together with L. amnigena and L. nimipressuralis, Enterobacter soli and Buttiauxella izardii (Figure 2). Moreover, the reconstructed phylogenetic tree based on partial rpoB (637 bp), atpD (642 bp), gyrB (742 bp) and infB (615 bp) gene sequences (Figure 3) indicated that strain ZJN belonged to Lelliottia genus, showing both 94.55% atpD gene sequence similarity, 92.94 and 96.23% rpoB gene sequence similarity, 88.27% and 89.08% gyrB gene sequence similarity and both 92.85% infB gene sequence similarity to L. amnigena and L. nimipressuralis, respectively (Table 1). Accumulatively, we assigned strain ZJN into the genus Lelliottia. Additionally, the Enterobacter aerogenes strains were clustered in the same clade as Klebsiella strains (Figure 3), suggesting that E. aerogenes should be reclassified as K. pneumoniae, consistent with the reports in Bergey’s Manual of Systematic Bacteriology [20] and Brady et al. [7].
Figure 3: Neighbor-joining tree showing the phylogenetic relationships of strain ZJN and phylogenetically related reference strains based on concatenated partial rpoB, atpD, gyrB and infB gene sequences. Bootstrap values based on 1000 resamplings are shown at branch nodes. Bar, 2% substitution rate per site.
Biochemical test on the strain
Further biochemical test showed that strain ZJN is Gram negative (Figure 4), facultatively anaerobic. Optimum temperature for growth is 30°C, grows at 37°C and 41°C, different from the L. amnigena and L. nimipressuralis (Table 2). Based on the biochemical characteristics in Biolog Gen database (Table 2) and the report by Brady et al. [7], this strain was identified to Lelliottia genus. However, it was positive for methyl red and oxidized D-sorbitol (Table 2), more closely related with L. amnigena than L. nimipressuralis and Leclercia adecarbxylata. In combination of the phylogenetic analysis and the results of biochemical test, we assigned strain ZJN as L. Amnigena.
| Characteristic | Leclercia adecarbxylata | Lelliottia amnigena | Lelliottia nimipressuralis | ZJN |
|---|---|---|---|---|
| Growth at temp | ||||
| 37°C | + | + | + | + |
| 41°C | ND | - | - | + |
| Motility | + | + | + | + |
| Growth in KCN | + | + | + | ND |
| Yellow pigment | + | - | - | - |
| Indole production | + | - | - | - |
| H2S production | - | - | - | - |
| Methyl red | + | V | - | + |
| Voges-Proskauer | - | + | + | + |
| Arginine dihydrolase | - | + | + | - |
| Ornithine decarboxylase | - | V | + | - |
| Lysine decarboxylase | - | - | - | - |
| Malonate | + | + | + | + |
| Citrate | - | + | + | + |
| D-Serine | - | V | V | - |
| Nitrate to nitrite | ND | + | + | + |
| Urease | ND | + | + | - |
| Gelatinase | - | - | - | - |
| β-galactosidase | + | + | + | + |
| Turanose | - | V | V | - |
| p-Hydroxy-phenylaceticacid | + | - | - | - |
| Fermentation of: | ||||
| Tween 40 | - | - | - | V |
| Tween 80 | - | - | - | V |
| D-Arabinose | + | + | + | + |
| Maltose | ND | + | + | + |
| D-Glucose | + | + | + | + |
| D-Lactose | + | + | + | + |
| D-Galactose | ND | + | + | + |
| L-Rhamnose | + | + | + | + |
| D-Mannose | ND | + | + | + |
| D-Fructose | ND | + | + | + |
| α-methyl-D-glucoside | - | V | ND | + |
| D-Trehalose | ND | + | + | + |
| D-Raffinose | V | + | + | V |
| Melibiose | + | + | + | + |
| D-Sucrose | V | V | - | - |
| D-Mannitol | + | + | + | + |
| meso-Inositol | - | - | - | V |
| D-Cellobiose (acid) | + | + | + | + |
| D-Cellobiose (gas) | + | + | + | + |
| D-Glycerol (gas) | ND | + | + | + |
| D-Sorbitol | V | V | - | + |
| Salicin | + | + | ND | + |
| D-Arabitol | + | - | - | V |
Table 2: Phenotypic features of strain ZJN and phylogenetically related species.
Pathogenicity test of the strain
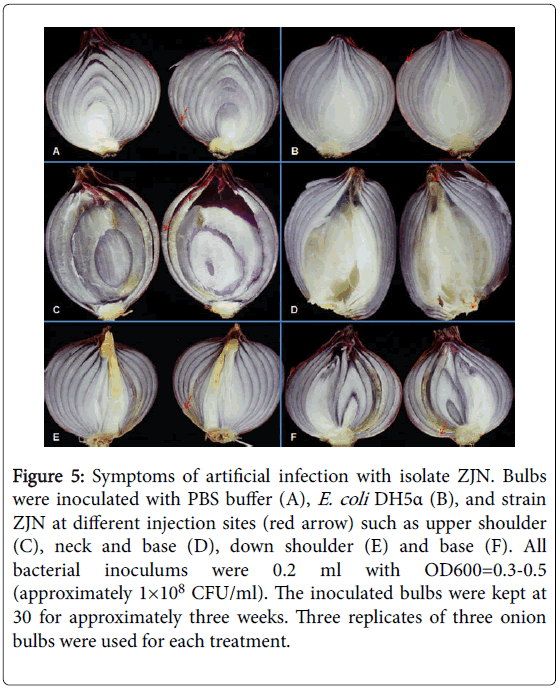
Subsequently, triplicates of onion bulbs were then injected at the neck, the base and the shoulder, with strain ZJN, E. coli DH5α and PBS buffer, respectively. After incubating at 30°C for 3 weeks, symptoms similar to the natural infections were observed on the ZJN inoculated bulbs, whereas, bulbs injected with PBS buffer and E. coli DH5α remained symptomless (Figure 5). However, different speeds and symptoms in disease development were indicated at different injection sites. When inoculating at the shoulder, the infected tissues of the bulbs became darker brown discoloration, dried out, collapsed and lesions extended through the site of inoculation, with intact peel (Figures 5A, 5C, 5D), whereas, when infecting at the neck or base, the internal fleshy scales of the bulbs were water soaking, with tissues maceration, necrosis and lesions extended outside (Figure 5B). Bacteria were reisolated from diseased tissues and further identified as L. amnigena ZJN by identical 16S rRNA gene sequence.
Figure 5: Symptoms of artificial infection with isolate ZJN. Bulbs were inoculated with PBS buffer (A), E. coli DH5a (B), and strain ZJN at different injection sites (red arrow) such as upper shoulder (C), neck and base (D), down shoulder (E) and base (F). All bacterial inoculums were 0.2 ml with OD600=0.3-0.5 (approximately 1×108 CFU/ml). The inoculated bulbs were kept at 30 for approximately three weeks. Three replicates of three onion bulbs were used for each treatment.
At this moment, we confirm that strain ZJN, which caused an Enterobacter bulb decay of onion, is L. amnigena according to the 16S rRNA gene sequence analysis, multilocus sequence analysis of rpoB, gyrB, infB and atpD gene sequences and biochemical features.
Discussion
Allium cepa L, also known as onion, is a popular vegetable and traditional medicine as a seasonal and storage crop. Whereas, onion bulb decay was recently reported as a storage disease caused by Enterobacteriaceae, including Enterobacter cloacae in Jordan, Australia, California, Colorado, Washington, New York and China [3,18,21-25] and Pantoea ananatis and Klebsiella pneumoniae in New York and China [26-28].
In this study, we identified another bacterial pathogen called Lelliottia amnigena which caused onion bulb decay in postharvest stage using a combination of taxonomic techniques such as morphological observations, biochemical test and MSLA analysis. Although the 16S rRNA gene sequence of strain ZJN showed the highest similarity with E. aerogenes (99.43%) (Table 1) and the concatenated partial rpoB, atpD, gyrB and infB gene sequence was most similar to that of Leclercia adecarboxylata (92.84%), strain ZJN was assigned in the same clade of Lelliottia in both phylogenetic trees (Figure 2 and Figure 3). However, there are some differences in the biochemical phenotypes between strain ZJN and L. amnigena and L. nimipressuralis, e.g. strain ZJN could grow at 41°C, while the other two species could not [7]; different from L. amnigena and L. nimipressuralis, strain ZJN was negative for arginine dihydrolase and urease (Table 2), suggesting that these characteristics may be variable in different strains. By comparing the other biochemical features, we found that strain ZJN was positive for methyl red and negative for ornithine decarboxylase, fully different with that of L. nimipressuralis, whereas, these features of L. amnigena are variable in different strains [7,29-31]. Accumulatively, we propose to assign this strain as L. amnigena.
After pathogenicity test of strain ZJN on healthy onion bulbs, we found that symptoms on different inoculation sites are different, which were darker and dried out at the shoulder with intact peel, whereas, water soaking, macerating and with lesions extended outside at the neck or base. We believed that the internal fleshy scales are more tender and watery, easier to be damaged and macerated, while the external scales are dry with little nutrient to feed the bacteria, leading little symptoms development outside.
Currently, Lelliottia is a newly built genus which includes just two species, e.g. L. nimipressuralis and L. amnigena, which were previously considered to genera Erwinia and Enterobacter, respectively. Brenner et al. proposed that Erwinia nimipressuralis should be transferred to the genus Enterobacter as E. nimipressuralis [29]. Whereas, Brady et al. proposed E. nimipressuralis and E. amnigenus to reclassify into L. nimipressuralis and L. amnigena respectively based on MLSA [7]. L. amnigena was multidrug-resistant Enterobacteriaceae strain [32,33], reported to infect heart transplant patient, causing septicemia and endophthalmitis [34-37]. Moreover, a L. amnigena isolate from agricultural soil could effectively reduce nitrate to ammonia [20] and was reported to have a good potential for use as an antifungal biocontrol agent. Vipul et al. isolated L. amnigenus from the sea dumps [38]. They found that the strain showed high chitinase production and produced proteases, inhibiting the growth of Fusarium sp. and Macrophomina phaseolina. Up till now, L. nimipressuralis were previously reported to naturally infect elm trees and exhibiting symptoms of wetwood disease on plants [6] and could cause wilt of pyracanth [39]. To our knowledge, this is the first report of L. amnigena naturally causing soft rot disease on plant.
Acknowlegements
This research was supported by the National Basic Research Program of China (973 Program, grant number 2015CB150600) and Natural Science Foundation of China (NSFC31201479). We are grateful to Songzhen Yang in Guangdong Institute of Microbiology for his great help in the the slide preparation for transmission electron microscopy of the bacteria.
References
- Wang CF, Praphat K, Xie GL, Zhu B, Li B (2008) Bacterial wilt of mulberry (Morus alba) caused by Enterobacter cloacae in China. Plant Dis 92: 483.
- Zhu B, Lou MM, Xie GL, Wang GF, Zhou Q, et al. (2011) Enterobacter mori sp. nov. associated with bacterial wilt on Morus alba L. Int J SystEvolMicrobiol 61: 2769-2774.
- Schroeder BK, Toit LJD (2009) First Report of Enterobacter cloacae causing onion bulb rot in thecolumbia basin of washingtonstate. Plant Dis 93: 323.
- Moreira SI, Dutra DC, Rodrigues AC, Oliveira JR, Dhingra OD, et al. (2013) Fungi and bacteria associated with post-harvest rot of ginger rhizomes in Espirito Santo, Brazil. Trop Plant Pathol 38: 218-226.
- Kado CI (2006) The prokaryotes: Erwiniaand related genera (3rd edn.), Springer 6: 443-450.
- Khodaygan P, Sedaghati E, Sherafati F (2012) Isolation ofEnterobacter nimipressuralis associated with bacterial wet wood from elm (Ulmus sp.) in Rafsanjan. Iran J Plant Pathol 47: 481-482.
- Brady C, Cleenwerck I, Venter S, Coutinho T, Vos PD (2013) Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. asLelliottia nimipressuraliscomb. nov. andLelliottia amnigena comb. nov., respectively, E. gergoviaeand E. pyrinusinto Pluralibacter gen. nov. as Pluralibactergergoviaecomb. nov. and Pluralibacterpyrinuscomb. nov., respectively, E. cowanii, E. radicincitans,E. oryzaeandE. arachidisinto Kosakonia gen. nov. as Kosakoniacowaniicomb. nov., Kosakoniaradicincitanscomb. nov., Kosakoniaoryzaecomb. nov. and Kosakoniaarachidiscomb. nov., respectively, and E. turicensis, E. helveticusandE.pulverisinto Cronobacteras Cronobacterzurichensis nom. nov., Cronobacterhelveticuscomb. nov. and Cronobacterpulveriscomb. nov., respectively, and emended description of the genera Enterobacterand Cronobacter. SystApplMicrobiol 36: 309-319.
- Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, et al. (1989) Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoeaagglomerans comb. nov. and description of Pantoeadispersasp. nov.Int J SystBacteriol 39: 337-345.
- Pavan ME, Franco RJ, Rodriguez JM, Gadaleta P, Abbott SL, et al. (2005) Phylogenetic relationships of the genus Kluyvera: transfer of Enterobacterintermedius Izard et al. 1980 to the genus Kluyvera as Kluyveraintermediacomb. nov. and reclassification of Kluyvera cochleaeas a later synonym of K. intermedia.Int J SystEvolMicrobiol 55: 437-442.
- Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, et al. (2008) Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobactersakazakiigen. nov., comb. nov.,Cronobactermalonaticussp. nov., Cronobacterturicensissp. nov., Cronobactermuytjensiisp. nov., Cronobacterdublinensissp. nov., Cronobactergenomospecies1, and of three subspecies,Cronobacterdublinensissubsp. dublinensis subsp. nov., Cronobacterdublinensissubsp. lausannensis subsp. nov. and Cronobacterdublinensissubsp. lactaridi subsp. nov.Int J SystEvolMicrobiol 58: 1442-1447.
- Kuhnert P, Korczak BM, Stephan R, Joosten H, Iversen C (2009) Phylogeny and prediction of genetic similarity of Cronobacterand related taxa by multilocus sequence analysis (MLSA). Int J Food Microbiol 136: 152-158.
- Gao J, Wang Y, Wang CW, Lu BH (2014) First report of bacterial root rot of ginseng caused by Pseudomonas aeruginosa in China. Plant Dis 98: 1577-1577.
- Coenye T, Falsent E, Vananneyt M, Hostef B, Govant JRW, et al. (1999) Classification of Alcaligenesfaecalis-like isolates from the environment and human clinical samples as Ralstoniagilardii sp. nov.Int J SystBacteriol 49: 405-413.
- Brady C, Cleenwerck I, Venter SN, Vancanneyt M, Swings J, et al. (2008) Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). SystApplMicrobiol 31: 447-460.
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, et al. (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J SystEvolMicrobiol62: 716-721.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. MolBiolEvol 28: 2731-2739.
- Grimont PAD, Grimont F (2005) Genus: Klebsiella, In: Volume Two: The Proteobacteria, Part B: The Gammaproteobacteria. In: Brenner DJ, Krieg NR, Staley JT (Edn.) Bergey’s Manual of Systematic Bacteriology, (2nd edn), Springer, New York, pp. 685-693.
- Schroeder BK, Toit LJD (2010) Effects of postharvest onion curing parameters on Enterobacter bulb decay in storage. Plant Dis 94: 1425-1430.
- Schroeder BK, Humann JL, Toit LJD (2012) Effects of postharvest onion curing parameters on the development of sour skin and slippery skin in storage. Plant Dis 96: 1548-1555.
- Fazzolari E, Mariotti A, Germon JC (1990) Nitrate reduction to ammonia: A dissimilatory process in Enterobacter amnigenus . Can J Microbiol 36: 779-785.
- Bishop AL (1990) Internal decay of onions caused by Enterobacter cloacae. Plant Dis 74: 692-694.
- Cother EJ, Vivienne D (1986) Bacteria associated with internal breakdown of onion bulbs and their possible role in disease expression. Plant Pathol 35: 329-336.
- Schwartz HF, Otto K (2000) First report of a bulb decay of onion by Enterobacter cloacae in Colorado. Plant Dis 84: 808.
- Zaid AM, Bonasera JM, Beer SV (2011) First report of Enterobacter bulb decay of onions caused by Enterobacter cloacae in New York. Plant Dis 95: 1581.
- Zhang Z, Nan Z (2013) Occurrence of lucerne seed-borne Enterobacter cloacaesprout decay in Gansu Province of China. Eur J Plant Pathol 135: 5-9.
- Carr EA, Bonasera JM, Zaid AM, Lorbeer JW, Beer SV (2010) First report of bulb disease of onion caused by Pantoeaananatisin New York. Plant Dis 94: 916.
- Carr EA, Zaid AM, Bonasera JM, Lorbeer JW, Beer SV (2013) Infection of onion leaves by Pantoeaananatis Leads to bulb infection. Plant Dis 97: 1524-1528.
- Liu SY, Lv MF, Gu YF, Zhou JN (2015) First report of bulb disease of onion caused by Klebsiellapneumonia in China. Plant Dis99: 1853.
- Brenner DJ, Mcwhorter AC, Kai A, Steigerwalt AG, Farmer JJ (1986) Enterobacter asburiaesp. nov, a new species found in clinical specimens, and reassignment of Erwiniadissolvensand Erwinianimipressuralis to the genus Enterobacter as Enterobacter dissolvenscomb. nov and Enterobacter nimipressuraliscomb. nov. J ClinMicrbiol 23: 1114-1120.
- Izard D, Gavini F, Trinel PA, Leclerc H (1981) Deoxyribonucleic acid relatedness between Enterobacter cloacaeand Enterobacter amnigenussp. nov.Int J SystEvolMicrobiol 31: 35-42.
- Tamura K, Sakazaki R, Kosako Y, Yoshizaki E (1986) Leclerciaadecarboxylatagen. nov., comb. nov., formerly known as Escherichia adecarboxylata. CurrMicrobiol 13: 179-184.
- Liu Z, Zhang Z, Yan H, Li J, Shi L (2015) Isolation and molecular characterization of multidrug-resistant Enterobacteriaceaestrains from pork and environmental samples in Xiamen, China. J Food Prot 78: 78-88.
- Murugaiyan J, Krueger K, Roesler U, Weinreich J, Schierack P (2015) Assessment of species and antimicrobial resistance among Enterobacteriaceae isolated from mallard duck faeces. Environ Monit Assess 187: 127.
- Bollet C, Elkouby A, Pietri P, Micco PD (1991) Isolation of Enterobacter amnigenus from a heart transplant recipient. Eur J ClinMicrobiol. Infect Dis 10: 1071-1073.
- Capdevila JA, Bisbe V, Gasser I, Zuazu J, Olivé T, et al. (1998) Enterobacter amnigenus. An unusual human pathogen. EnfermInfeccMicrCl 16: 364-366.
- Jan D, Berlie C, Babin G (1999) Fatal posttransfusionEnterobacter amnigenus septicemia. Presse Med 28: 965.
- Westerfeld C, Papaliodis G, Behlau I, Durand M, Sobrin L (2009) Enterobacter amnigenusendophthalmitis. Retin Cases Brief Rep 3: 409-411.
- Vipul G, Tejas C, Pranav V, Chhatpar HS (2004) Isolation and identification of marine chitinolytic bacteria and their potential in antifungal biocontrol. Indian J ExpBiol 42: 715-720.
- Sartori JF, Fucikovsky L (1976) Erwinianimipressuralis, causal agent of pyracanth(PyracanthakoidzumiiReh.) wilt. Rev LatinoamMicrobiol 18: 185-187.
- Zelalem T, Douglas RT, Franklin RC (1997) Leclerciaadecarboxylatainfections: case report and review.Clin Infect Dis 25: 79-81.
Copyright: © 2016 Liu S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.