Journal of Agricultural Science and Food Research
Open Access
ISSN: 2593-9173
ISSN: 2593-9173
Research Article - (2024)Volume 15, Issue 1
Rice is mainly a semi-aquatic plant, an adequate amount of water is necessary for the better development and growth of rice crops. But due to the scarcity of water in rice-growing countries, they faced problems like drought stress and other abiotic stresses. The research is conducted to analyze the root system and root morphology of rice plants because the rice is drought tolerant and directly correlated with the root system. The study is designed for rice root traits analysis i.e., genotypic and phenotypic. The hundred diverse genotypes of rice with three different groups viz. wild type, the cultivated type and germplasm rice were grown under the rhizotron conditions. The 157 SSR markers were used for studying the population of rice and genotypic data were generated. The root was scanned by root scanner after 45 Days After Sowing (DAS) and the parameters taken for the considerations are Root Length (RL), Total Root Length (TRL), Average Root Diameter (ARD), Surface Area (SA), Root Weight Fresh (RWF), Root Weight Dry (RWD) and Root Volume (RV) etc. Phenotypic data was recorded. Six markers were found associated with the root traits. Reference Marker (RM-408) on chromosome 8 is associated with the TRL is, also associated with the fresh root weight. RM-236 and RM-408 on chromosome-8 is associated with root surface area, NOD-1 and NOD-3 on chromosome-8 is associated with the dry root weight. RM-505 on chromosome-7 is associated with the average root diameter, RM-1 on chromosome-1 is also associated with the root surface area these markers show the marker trait associations in rice. Genotypes Wilt Resistant (WR-41), Rashtriya Krishi Vikas Yojana-104 (RKVY-104), Bamleshwari, Nagina-22 and Conventional Tillage (CT-9993) having good root length, root volume, root diameter etc. This study will help in future for selection and use of donor genotype of rice for drought tolerance.
Rice; Drought; Root traits; Maker-trait association
RL: Root Length; TRL: Total Root Length; ARD: Average Root Diameter; SA: Surface Area; RWF: Root Weight Fresh; RWD: Root Weight Dry; RV: Root Volume; NOD: Nitric Oxide Dismutase; RM: Rice Microsatellite; QTLs: Quantitative Trait Locus; DNA: Deoxyribonucleic Acid
The rice root system is essential for the uptake of water, nutrients and structural support. Strong shoots are a requirement for higher energy utilization efficiency and more photosynthesis. Deep, thick and branched roots are responsible for better survival under adverse climatic conditions. Rice transplanting was damaged by global water shortage, strong roots are conducive to rice growth in middle to late periods and stable yield [1].
New research and discoveries of genes related to traits and shoot traits are important for developing rice seedlings with survivability and tolerance to abiotic stress. In rice root system traits like RL, RW, RLV and RT is important for better survival under severe conditions, like water and nutrient deficiency, however, the measurement of these difficult traits was considered the most difficult [2]. Root trait is a quantitative trait controlled by many genes responsible for the drought, each with a small genetic effect [3].
Selection and breeding for desirable root traits associated with drought tolerance have been practiced in rice and the differential response of rice genotypes to drought has been related to root system characters [4,5]. Plant response to drought stress is one of the most complex biological processes and it involves numerous physiological, cellular and molecular levels. Many genes have been identified to be involved in the response to drought stress in plants [6]. Under non-flooded conditions, the inefficient water and nutrient supply due to the poor root structure from puddled transplanted rice need to be addressed through trait and varietal developmental programs. The root system architecture of plants is dependent on nutrient and water availability and uptake signaling. There is an urgent need to decipher an appropriate plant and root system architecture for improving nutrient uptake, grain yield and adaptability for better crop improvement [7]. To study, genetic control for root development, association mapping is very helpful. The genetic dissection of the root system of upland rice by using QTL mapping, to investigate the relationship between different root traits of rice and drought resistance under different water stress conditions and also identification of QTL responsible for root traits under different developmental stages. Mapping Quantitative Trait Loci (QTLs) and association mapping for root traits and the Marker-Assisted Breeding program (MAB) help to overcome the water scarcity problems.
Plant materials
To conduct this experiment, we have taken 100 different rice genotypes containing landraces and germplasm and cultivated those varieties that were grown in rhizotron condition in Plant Molecular Biology and Biotechnology (PMBB) rainout for 45 days then, root material was used for root scanning, we have used these genotypes because these rice genotypes contain standard varieties as well as hybrids with different types of root morphology (Table 1).
| S/no. | Genotypes name | S/no. | Genotypes name | S/no. | Genotypes name | S/no. | Genotypes name |
|---|---|---|---|---|---|---|---|
| 1 | Annada | 26 | IBD-1 | 51 | Sehra dabri | 76 | Chhind, Guchchhi |
| 2 | Abhaya | 27 | Danteshwari | 52 | Tarunbhog | 77 | Nariyal, Phool |
| 3 | Azucena | 28 | Poornima | 53 | Chapti gurmatia | 78 | Ama, Ruthi |
| 4 | ARB6 | 29 | Sahabhagi, Dhan | 54 | Kalam gurmatia (3053) | 79 | Khajur, Jhopa |
| 5 | Bamleshwari | 30 | MTU 10106 | 55 | Srikamal | 80 | Naykain, Jhaba |
| 6 | Buddha | 31 | Aganni | 56 | Elayachi | 81 | Ram, Laxman |
| 7 | Bakal | 32 | Karma masuri | 57 | Bisni-I | 82 | Do, Dana |
| 8 | Bhatapool | 33 | Safri 17 | 58 | CGZR-1 | 83 | Pankhi |
| 9 | Batroo | 34 | Dubraj | 59 | Basmati 370 | 84 | Sua, Pankhi |
| 10 | CT 9993 | 35 | BPT 5204 (Improved) | 60 | Kalamata | 85 | Maina, Gali |
| 11 | Cross 116 | 36 | Jitpiti | 61 | Jhilli | 86 | Parewa |
| 12 | Deshi lal, Dhan | 37 | PR 122 | 62 | Ramjiyawan | 87 | Chini, Kapoor |
| 13 | Dagad, Deshi | 38 | WR 32 | 63 | RKVY-104 | 88 | Pakshi raj |
| 14 | IR 62266 | 39 | WR 36 | 64 | BAM 5926 | 89 | Surmatia |
| 15 | IR 36 | 40 | WR 41 | 65 | Moroberekan | 90 | Parmal |
| 16 | IR 42253 | 41 | WR 99 | 66 | Naginna 22 | 91 | Dokra, Mechha |
| 17 | IR 64 | 42 | WR 116 | 67 | GP-145-42 | 92 | Jai gundi |
| 18 | IR 55419-04 | 43 | E-1702 | 68 | GP-145-43 | 93 | Khatia, Pati |
| 19 | Kranti | 44 | Maheshwari | 69 | Kalokuchi | 94 | Tulsi, Prasad |
| 20 | Lalmatia | 45 | Durgeshwari | 70 | Shenong | 95 | CR-1014 |
| 21 | Laloo-14 | 46 | Shymala | 71 | IR-55419 | 96 | Anterved |
| 22 | Mahamaya | 47 | Rajeshwari | 72 | R-RF-75 | 97 | Layacha |
| 23 | Samleshwari | 48 | Chandrahasini | 73 | Kadam, Phool | 98 | Tulsi, Manjari |
| 24 | Swarna | 49 | Jaldubi | 74 | Ama jhopa | 99 | Kera, Ghul |
| 25 | Vandana | 50 | Indira sugandhit dhan1 | 75 | Koudi dhull | 100 | Kubri mohar |
Note: IDB: International Dairy and Beef; CZR: Chhattisgarh Zinc Rice; PR: Protease.
Table 1: Details of 100 rice genotypes used in the study.
Rhizotron study
Root characters were studied in rhizotron condition. At the end of the experiment, roots were washed and then scanned for analysis of root traits using software Win Rhizo viz. root length, root volume, surface area, root diameter was used for QTLs analysis (Figure 1).

Figure 1: Rice Genotypes under rhizotron condition.
Root scanning of selected germplasm rice lines: The roots of each germplasm were used for scanning. The roots of three plants from each germplasm line/variety were used for root scanning which given the detailed information about all root parameters including root length, root volume, root diameter and root surface area etc. The root scanning was carried out by using root scanner machine Epson perfection V700/V750, 3.81 Version, Win Rhizo Reg 2009, the data was recorded automatically in the computer for different root parameters including root length, average root diameter, root volume and root surface area. Following procedure was used for root scanning (Table 2).
| Stock | Aliquot |
|---|---|
| DNA 25 ng/ml | 2.0 ml |
| dNTPs (2.0 mM) | 4.0 ml |
| Primer (10 mM) (F+R) | 1.2 ml |
| PCR reaction buffer (2X) | 10.0 ml |
| Taq DNA polymerase | 0.5 ml |
| Sterile distilled H20 | 2.3 ml |
| Total | 20 µl |
Note: DNA: Deoxyribose Nucleic Acid; dNTPs: Deoxynucleotide Triphosphates; PCR: Polymerase Chain Reaction.
Table 2: Reaction mixture (20 µl) for Polymerase Chain Reaction (PCR) with gene specific 157 primers.
Acquiring washed roots: The first step is acquiring washed roots. This can be the most difficult and laborious step in the experiment if plants are grown in solid medium. For root scanning, roots washed with tap water for 2-3 times gently so that fine roots would not damage to remove soil completely and the roots were preserved in the spirit solution (25%) in falcon tubes for root scanning. The procedure was conducted very cautiously to prevent supplementary root damage and losses. Unwanted debris and dead roots were removed from vital roots.
Preparing roots for scanning: The 3-4 acrylic trays were washed with water completely, then by using tissue paper wipe out water, clean properly and dried completely. The tray filled with water (3/4th portion of the tray so that scanning will be cleared) and the roots were placed in a tray. Then the roots were floated in water in the acrylic trays in the scanner, so we need to arrange these roots to reduce the overlapping and crossing of roots for obtaining accurate image and other details. Plastic forceps were used to arrange the roots in specific manner, this is so delicate work, enough lighting and steady hands are helpful for this work.
Scanning of roots: For obtaining best results, Win Rhizo Reg 2009 with an approved scanner used for root scanning, which allow the roots to be light from above and below while root being scanned. This is an important feature (called “dual scan” in Regent’s documentation), which reduces shadows on the root image. The reagent positioning system allows the trays to be consistently placed thus obviating the need to preview each scan. Optimum scanning resolution depends on the type of samples. Generally, roots scanned at 600 dpi in 10 × 15 cm trays. RL analysis was carried out with grayscale images.
The right threshold value is important: Analysis results can be sensitive to the threshold parameters used. Win Rhizo can automatically set these, one you may manually tweak them from time to time. The color traces on the root indicate where roots have been detected.
Analyzing scanned images: For image analysis we need to select the image area or region of interest and then it is analyzed. After image has been scanned, the software uses threshold to determine which one is the root and which is not root or other debris. After few seconds, the analysis was completed and roots found by Win Rhizo were identified by colored line in image. The colors used for drawing them are coded according to root diameter. Portions of the image can be excluded from analysis if necessary and there are basic tools available if any minor image editing is required.
Saving of measured data: The last step of root analysis was data saving Win Rhizo knows when data was easily recordable by many programs including spreadsheet style like excel. Image and their analysis were also saved to file for later validation, reanalysis or for visualization in other software programs. Rhizotron observation for plant production and root trait. The data was recorded after 45 days after sowing in which TRL, root diameter, root volume and surface area will be recorded by using root scanner (Figure 2).

Figure 2: Rhizotron observation for root trait.
• Root length: The total RL was recorded in cm unit from the point of attachment to shoot to the tip of root using scale.
• Total root length: The total RL was measured by root scanner in millimeter.
• Root weight (fresh): The fresh root weight was measured by using the weighing balance in grams.
• Root weight (dry): The dry root weight was measured on weighing balance after oven drying for 24 hrs.
• Total root diameter: The root diameter was measured by root scanner after root scanning in mm.
• Root surface area: The root surface area was measured by root scanner in cm².
• Root volume: The root volume was measured by root scanner in cm³.
Genotyping of the selected rice germplasm
Genomic DNA isolation: Total rice germplasm DNA was extracted from hundred rice germplasm lines by modified Cetyltrimethylammonium Bromide (CTAB) protocol for DNA extraction and used for Polymerase Chain Reaction (PCR) amplification to test the genes for root traits using 157 different molecular markers [8].
Genomic data was taken from Rice Genome Project which was obtained by following protocol for isolation of genomic DNA. The leaves from each plant was cut into small pieces with the help of sterile scissor. Once the sample is prepared 200 μl Ethoxybenzoic Acid (EBA) and 80 μl Sodium Dodecyl Sulfate (SDS) was added and sample was crushed using tissue lyzer (Mo Bio Laboratories ltd. Pawerlyzer™ 24). After crushing the samples were vortex and incubated in water bath at 65°C for 10 min. To this added 600 μl of chloroform: Isoamyl alcohol (24:1) and gently shake the mixture by inversion for 15 min. Centrifuged at 13,000 rpm for 10 min to separate the phase and transferred the upper phase to new tube. Repeat the chloroform extraction (step 4) one more time. Added 2/3rd volume of pre-chilled isopropanol and incubated at room temperature -20°C for 30 min or longer until DNA was precipitated. Centrifuge for 10 min at 13,000 rpm to collect DNA pellet. Washed the DNA pellet with 70% ethanol and air dry for 10 min. Re-suspend the DNA pallet in 200-500 μl of TE buffer and allowed the pallet to dissolve. Added 2-5 μl of Ribonuclease (RNase) (10 mg/ml) and incubated at 37°C for 30 min to this added 1/10th volume of 3 Molar (M) sodium acetate and 2 M volume of pre-chilled absolute ethanol. Mix gently and incubate at room temperature for 30 minutes if a large amount of DNA is precipitated, there was no need to incubate for 30 minutes. Centrifuged the DNA pellet for 5 minutes at 13,000 rpm. Prolonged centrifugation yields hard DNA pellet which was difficult to re-suspend in Tris-EDTA (TE) buffer. Then added 1 ml of 70% ethanol and washed the DNA pellet. Centrifuged for 1 minute at 13,000 rpm. Discard the 70% alcohol and placed the tube upside down on paper towel to get rid of excess of ethanol. Re-suspended the pellet in 100-200 μl of TE buffer and incubate overnight; the volume of TE buffer must be depended on size of DNA pellet. Centrifuged at 13,000 rpm for 5 minutes and transfer supernatant to new 1.5 ml of micro centrifuge tube. Stored at -20°C until use.
Genomic DNA quantification: The DNA samples were quantified by using Nano Drop (ND) spectrophotometer thermo scientific (ND 100). DNA concentration values in ng/ μl were obtained on the basis of absorbance at 260 nm.
Protocol: The sampling arm’s upper and lower pedestals were cleaned by wiping it with a soft and sterile tissue paper. Water sample (1 μl) was loaded onto the sampling arm to clean the sample column and to initialize spectrometer ready for use. 1 μl TE buffer (blank sample) was loaded for initial blank measurement and both the pedestals were wiped after the blank value was obtained. 1 μl of DNA samples were loaded sequentially and the readings on concentration of DNA in ng/ μl were stored as image file with sample details and spectral measurements.
Polymerase Chain Reaction (PCR) standardized protocol
The PCR protocol for genotyping was standardized as follows.
Add required reagents or master-mix and template to PCR tubes. Mix and centrifuge. Amplify per thermo cycler and primer parameters. Evaluate amplified DNA by agarose gel electrophoresis followed by ethidium bromide staining. This is a basic PCR protocol using Taq DNA polymerase. PCR in the absence of exogenously added DNA was used as negative control to check the DNA free status of reagents and solutions. PCR with positive control DNA was used to check the completeness of the PCR mixture (to check the quantity of essential components including MgCl2 in the cocktail). Negative and positive controls were used to check for spurious background bands and reaction specificity and to identify PCR parameters (includes annealing and number of cycles) that were suitable for amplifying expected products.
PCR amplification: The PCR for amplification was prepared as follows in. The reaction mixture was given a momentary spin for throughout mixing of the cocktail components. Then 0.20 ml PCR tubes were loaded in a thermal cycler. The reaction in thermal cycler was programmed as shown in Table 3.
| Profile 1 | 95°C for 5 minutes | Initial denaturation |
| Profile 2 | 95°C for 1 minute | Denaturation |
| Profile 3 | 55°C for 1 minute | Annealing |
| Profile 4 | 72°C for 1 minute | Extension |
| Profile 5 | 72°C for 7 minutes | Final extension |
| Profile 6 | 4°C | Hold the samples |
Table 3: Temperature profile of thermal cycler.
Polyacrylamide Gel Electrophoresis (PAGE)
Seven per cent polyacrylamide gels (vertical) were used for better separation and visualization of less size PCR amplified products, since polyacrylamide gels have better resolution for amplified products. Gels were casted in Columbia Broadcasting System (CBS)-scientific electrophoresis unit. Glass plates were prepared before making the gel solution. Both outer and inner notched glass plates were cleaned thoroughly with tap water, detergent and then with 70% ethanol.
Assembling and pouring the gel: Gasket was fixed to the three sides of the outer plate without notches. Spacers of 1.5 mm thickness were placed along the sides by just attaching the gasket of outer plate. Later, notched plate was kept on the outer plate so that, spacers were between the two plates. Clamps were put on the three sides of the plates leaving notch side of the unit. It was checked with water to find any leakage. For casting, gel was prepared just prior to pouring. To prepare the gel, 65 ml of 7% PAGE solution, 700 μl of ammonium per sulphate, 70 μl of N, N, N”, N”-tetra methylene diamine was added to initiate the polymerization process. The content was mixed by swirling, but bubbles were avoided. Before pouring, assembly was kept on the bench top so that it made 45° angle with bench top. Then gel solution was poured from notch side and air bubbles were avoided. Comb of 1.5 mm thickness (60 wells) was inserted with tooth side in the gel. Later, assembly was kept for polymerization for 20-30 minutes. After polymerization process, gasket and clamps were removed and kept in electrophoresis unit with electrophoresis unit clamps were notch side faced inner side of the unit. 1X Tris Borate- EDTA (TBE) was poured in the upper tank of the unit and the rest was poured in the bottom chamber. Comb was removed with care so that it did not disturb the wells formed. 3 μl loading dye was added to 10 μl PCR products. Finally, 5 μl of each sample were loaded into the wells for facilitating the sizing of the various alleles ladder (50 bp) was loaded in the first well. Electrophoresis was done at 180 volts till the dye reach the bottom of the gel.
Gel staining: Measure 1 g of agarose. Mix agarose powder with 100 mL 1X TAE in a microwavable flask. See TAE recipe.
Note: Make sure to use the same buffer as the one in the gel box (do not mix different buffers and do not use water). Microwave for 1-3 min until the agarose is completely dissolved (but do not over boil the solution, as some of the buffer will evaporate and thus alter the final percentage of agarose in the gel, many people prefer to microwave in pulses, swirling the flask occasionally as the solution heats up). Let agarose solution cool down to about 50°C (about when you can comfortably keep your hand on the flask), about 5 min. Add Ethidium Bromide (EtBr) to a final concentration of approximately 0.2-0.5 μg/mL (usually about 2-3 μl of lab stock solution per 100 mL gel). EtBr binds to the DNA and allows you to visualize the DNA under Ultra-Violet (UV) light.
Caution: EtBr is a known mutagen. Wear a lab coat, eye protection and gloves when working with this chemical. Pour the agarose into a gel tray with the well comb in place. Place newly poured gel at 4°C for 10-15 min or let sit at room temperature for 20-30 min, until it has completely solidified.
Loading samples and running an agarose gel: Add loading buffer to each of your DNA samples. Once solidified, place the agarose gel into the gel box (electrophoresis unit). Fill gel box with 1X Tris- Acetate-EDTA (TAE) or TBE until the gel is covered. Carefully load a molecular weight ladder into the first lane of the gel. Carefully load your samples into the additional wells of the gel. Run the gel at 80-150 V until the dye line is approximately 75-80% of the way down the gel. A typical run time is about 1-1.5 hours, depending on the gel concentration and voltage. Turn OFF power, disconnect the electrodes from the power source and then carefully remove the gel from the gel box.
Optional: If you did not add EtBr to the gel and buffer, place the gel into a container filled with 100 mL of TAE running buffer and 5 μl of EtBr, place on a rocker for 20-30 min, replace EtBr solution with water and distain for 5 min. Using any device that has UV light, visualize your DNA fragments. The fragments of DNA are usually referred to as ‘bands’ due to their appearance on the gel.
Analyzing your gel: Using the DNA ladder in the first lane as a guide (the manufacturer's instruction will tell you the size of each band), you can infer the size of the DNA in your sample lanes. For more details on doing diagnostic digests and how to interpret them please see the diagnostic digest page.
Purifying DNA from your gel: If you are conducting certain procedures, such as molecular cloning, you will need to purify the DNA away from the agarose gel. For instructions on how to do this, visit the gel purification page.
Visualization of amplified PCR products through PAGE: The gel was placed in the Gel Documentation System (GDS) for visualizing the amplified PCR products.
Genotyping: The genotypic study was done by performing the DNA isolation, Polyacrylamide Gel Electrophoresis (PAGE), Polymerase Chain Reaction (PCR) and gel staining under Rice Genome Project (RGP).
DNA isolation: Total rice germplasm DNA was extracted from hundred rice germplasm lines by Modified CTAB protocol for DNA extraction and used for PCR amplification to test the genes for root traits using 157 different molecular markers [9].
The PCR protocol for genotyping was standardized as follows.
Add required reagents or master-mix and template to PCR tubes. Mix and centrifuge. Amplify per thermo cycler and primer parameters. Evaluate amplified DNA by agarose gel electrophoresis followed by ethidium bromide staining. This is a basic PCR protocol using Taq DNA polymerase. PCR in the absence of exogenously added DNA was used as negative control to check the DNA free status of reagents and solutions. PCR with positive control DNA was used to check the completeness of the PCR mixture (to check the quantity of essential components including MgCl2 in the cocktail). Negative and positive controls were used to check for spurious background bands and reaction specificity and to identify PCR parameters (includes annealing and number of cycles) that were suitable for amplifying expected products.
Statistical analysis of primer data: The statistical analysis of primer data and the association mapping data were analyzed by using the software’s like Structure for finding the diversity among the rice population. Tassel software is used for marker-trait association analysis of rice genotype for root traits. In structure analysis the parameters were set as number of individual 100, number of loci 120, number of ploidies 2, missing value represented as-999, length of burning period 1000.
Diversity in rice genotype
We have performed the structure program to find out the diversity among the rice genotype they showing K=3 means the tree different types of rice genotypes these are the wild type, cultivated type and germplasm rice. Out of 100 genotypes, 85 genotypes (Sr.no. 1-66, 70, 72, 74, 76-82, 84, 86, 87, 89, 90, 93-96) comes under population 1, 13 genotypes (Sr.no. 67-69, 71, 73, 75, 83, 85, 88, 91, 92, 97, 98) comes under population 2 and 2 genotypes (Sr.no. 99, 100) comes under population 3. The diversity between the rice is desirable for selection of better rice genotype to perform the experiments. In this study we have performed the structure harvester and obtained results showing the diversity among the rice genotypes. The seven genotypes showing the pure line, remaining 93 genotypes of rice showing the admixture (Figures 3-5).
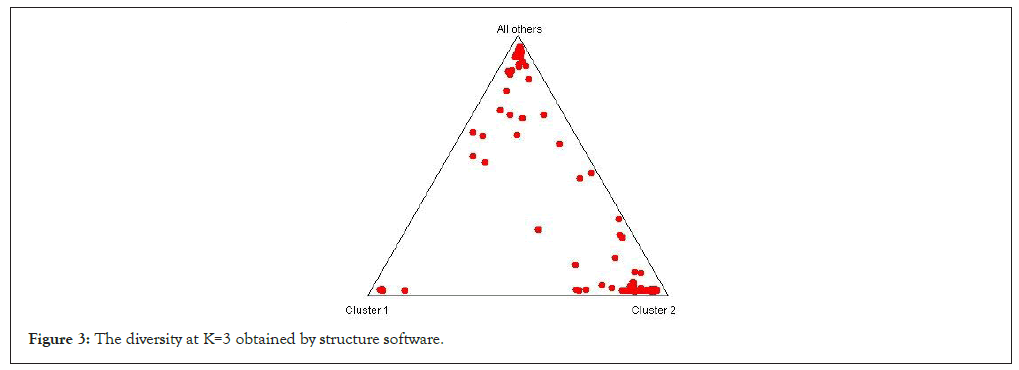
Figure 3: The diversity at K=3 obtained by structure software.
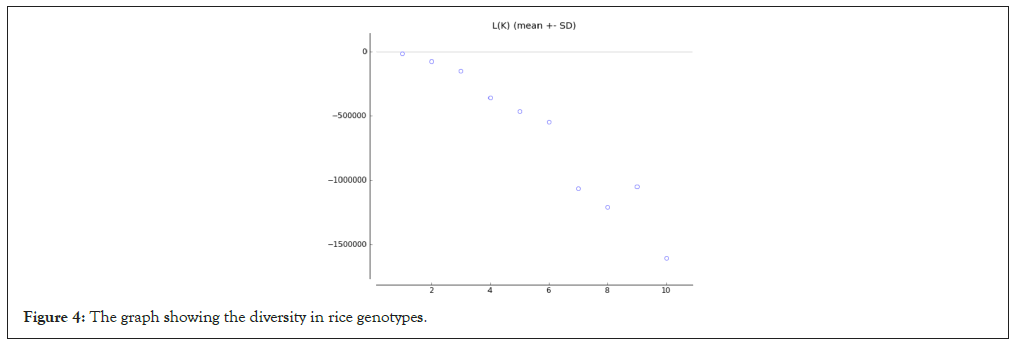
Figure 4: The graph showing the diversity in rice genotypes.
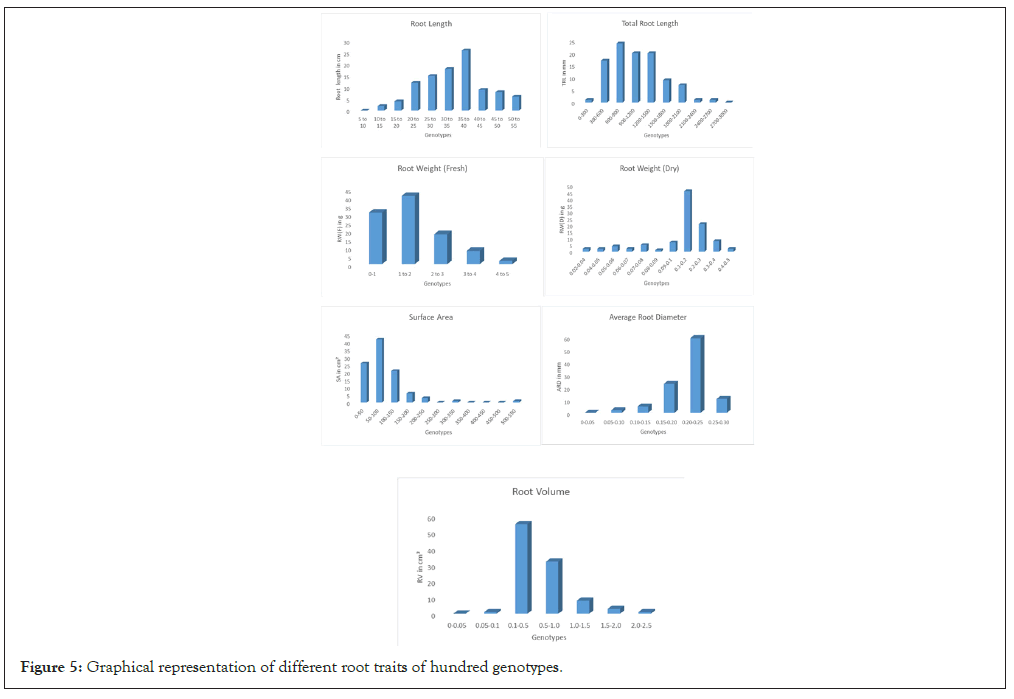
Figure 5: Graphical representation of different root traits of hundred genotypes.
Root traits
The root system is a one of the most important system in case of drought in this study some genotypes having the strong root morphology and genetic structure which is helpful in response to water stress. The root volume was considered as important traits because more the root volume it will absorb more amount of water from surrounding soil. RL shows the penetrance of roots in soil, it was need to take under the consideration. The highest RL was measured with scale was 55.42 cm in WR-41 genotype and the lowest RL was observed in Moreberekan which was 11.37 cm with mean of 37.47 cm and the high range of variation was observed among the 100-rice genotypes. The fresh root weight was recorded higher in the WR-41 was 4.53 g and the lowest was recorded in Moreberekan which was 0.18 g this is comparatively very less than WR-41. Average root weight is 1.49 g the earlier founding showing decreasing in RL and fresh root weight and dry root weight in BRRI Dhan rice varieties under stress condition. The root weight was recorded after oven drying for 24 hrs. The highest amount of dry weight was found in RKVY-104 was 0.46 g and the lowest was recorded in the WR-32 was 0.02 g with mean of 0.15 g. The higher total RL was observed in CT-9993 which was 2702.16 cm and the lowest was observed in the WR-36 which was 305.06 cm with mean of 745.86 cm the population showing the enormous diversity among each other. The root surface area is one of important root trait for drought and, the large surface area was recorded in Bamleshwari was 523 cm² and smaller surface area was observed in the WR-36 which was recorded 17.41 cm² with mean of 74.85 cm2. In root traits mainly; ARD and long specific root length, RL density helps in maintaining plant productivity because roots play an important role in absorbing water and nutrients required for plant growth [10]. The ARD was recorded highest in Nagina-22 that is 0.28 mm and the lowest was observed in Moreberekan was 0.07 mm with mean of 0.23 mm and representing the variation among the population. Root volume in rice is important because of water absorption capacity and root volume was highest in the RKVY- 104 was 2.04 cmꝪ and lowest in WR-36 which was recorded 0.08 cm³. Average root volume is 0.33 cm³. They investigated that the phenotypic variation of root traits, selected parents were different (P<0.05) for root volume and root dry weight. Out of 100 genotypes, genotypes WR-41, RKVY-104, Bamleshwari, Nagina-22 and CT-9993 having good root length, root volume, root diameter etc. Frequency distribution of RL showing normal distribution and other trait showing nearly normal distribution [11-13] (Figure 6).
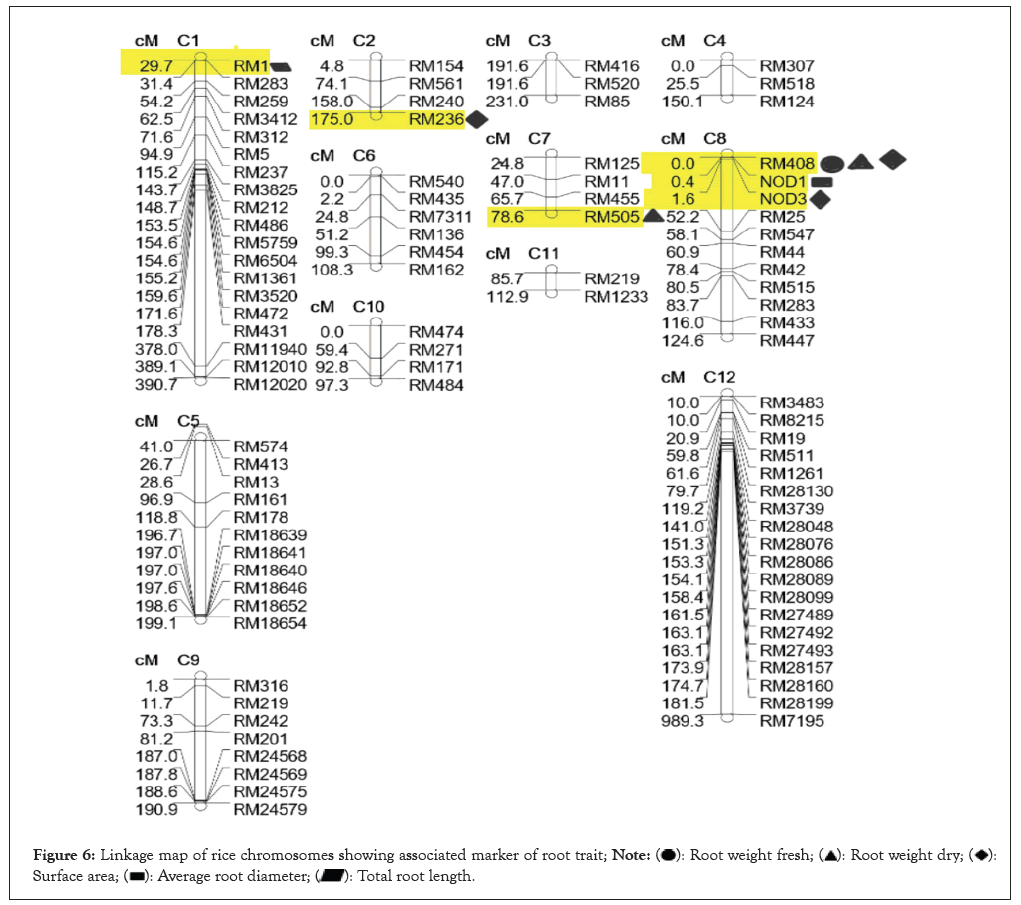
Figure 6: Linkage map of rice chromosomes showing associated marker of root trait; 
Marker-trait association
In this study we have found some marker associated with the various traits. Out of 157 SSR markers, 151 showing single banding (monomorphic) and (RM-3739, RM-505, RM-24569, RM-27489, RM-18652 and GW-8) showing multiple bands (polymorphic). Many researchers already found many markers that are responsible for different rice root traits such as the genomic regions on chromosomes 1, 4, 8 and 9 responsible for grain yield under stressed condition, DRO 3 which is new QTL for RGA on chromosome 7 in rice. Many droughts responsive QTLs also found for grain yield under drought stress, 9 QTLs simultaneously associated with more than two traits with common loci having effects on drought-related traits [14-17].
General Linear Model (GLM)
In this study the 157 SSR markers were analyzed for 100 rice accessions and out of which six markers were identified for various morphological root traits. In the molecular distinguishes among rice landraces in central India study showed that the marker RM-408. We have found marker RM-408 on chromosome-8 is associated with the Total RL (TRL) at >0.05 level of significance. Marker RM-505 were found associated with the RW(D) at <0.5 level of significance on chromosome-7. Marker RM-408 on chromosome-8 found in association with the fresh root weight at >0.05 level of significance, RM-408 on chromosome-8 and RM-236 on chromosome-2 is associated with the root surface area at >0.5 level of significance, also NOD-3 located on chromosome-8 is associated with surface area at <0.5. Marker NOD-1 on chromosome-8 associated with the ARD these markers not found earlier. This analysis will help to find out the drought related QTLs and markers. RM-1 located on chromosome-1 found associated with TRL at >0.5% level of significance (Table 4).
| Trait | Marker | Locus | Locus pos | Marker F | Marker p | Perm p | Marker R2 | Marker | Marker MS | Error | Error MS | Mode lDF | Mode lMS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RW(F) | RM408 | UK | 19 | 13.92 | 3.58E-04 | 0.038 | 0.1451695 | 1 | 2928952 | 79 | 210485.68 | 4 | 886927.57 |
| RW(D) | RM505 | UK | 34 | 16.48 | 1.13E-04 | 0.082 | 0.160493 | 1 | 0.02092624 | 81 | 0.0012699 | 4 | 0.0068823 |
| RW(D) | RM408 | UK | 19 | 12.77 | 6.03E-04 | 0.104 | 0.1347784 | 1 | 9.60389749 | 79 | 0.7518519 | 4 | 2.9651552 |
| TRL | RM1 | UK | 0 | 11.05 | 1.79E-09 | 0.14 | 0.507426 | 7 | 30056.6424 | 74 | 2720.8429 | 10 | 21329.248 |
| SA | RM408 | UK | 19 | 14.25 | 3.09E-04 | 0.419 | 0.15122 | 1 | 62191.1682 | 79 | 4365.296 | 4 | 16601.148 |
| SA | RM236 | UK | 113 | 12.97 | 5.92E-04 | 0.461 | 0.1542392 | 1 | 61157.2066 | 69 | 4715.268 | 4 | 17788.816 |
| SA | NOD3 | UK | 70 | 7.3 | 2.22E-04 | 0.587 | 0.2131664 | 3 | 0.32847324 | 79 | 0.0450157 | 6 | 0.1777549 |
| ARD | NOD1 | UK | 69 | 6.28 | 7.52E-04 | 0.687 | 0.2001895 | 3 | 0.3042133 | 73 | 0.0484554 | 6 | 0.1702722 |
Note: NOD: Nitric Oxide Dismutase; RM: Rice Microsatellite; UK: Unknown; ARD: Average Root Diameter; RW(D): Root Weight (Dry); TRL: Total Root Length; RW(F): Root Weight (Fresh); SA: Surface Area: MS: Microsoft; IDF: Intelligent Debris Free; IMS: Information Management System.
Table 4: Tassel results obtained from General Linear Model (GLM) model.
Mixed Linear Model (MLM)
In the MLM model of tassel analysis no significant results were obtained except marker RM-1 on chromosome-1 found to be associated with the root surface area at <5% level of significance. The marker locus is not identified and genetic variance was 0.02514 which is non-significant. The other markers were not found associated with the traits like RL, ARD, TRL, SA, RV and root fresh and dry weights. The high level of accuracy can be obtained in the MLM model. In this MLM study, we had found only one marker which is not satisfactory result but after doing multiple time analysis we found same consequences (Table 5).
| Trait | Marker | Locus | Site | Df | F | P | Error df | marker R² | Genetic var | Residual var | 2Ln Likelihood |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RL | None | - | - | 0 | None | None | 92 | None | 14.4905 | 83.341745 | 666.5536571 |
| RW(F) | None | - | - | 0 | None | None | 92 | None | 8.48E-06 | 0.8475566 | 244.2018134 |
| RW(D) | None | - | - | 0 | None | None | 92 | None | 0.0023224 | 0.0483156 | -13.81184712 |
| TRL | None | - | - | 0 | None | None | 92 | None | 34078.494 | 216805.84 | 1382.096151 |
| SA | None | - | - | 0 | None | None | 92 | None | 1081.2148 | 3979.5468 | 1023.282101 |
| SA | RM-1 | UK | 0 | 7 | 11.75 | 2.66E-10 | 84 | 0.47945 | 0.02514 | 2514.0037 | 886.409663 |
| ARD | None | - | - | 0 | None | None | 92 | None | 3.19E-05 | 0.0014816 | -333.0749805 |
| RV | None | - | - | 0 | None | None | 92 | None | 0.0055681 | 0.1416294 | 82.84408594 |
Note: UK: Unknown; RL: Root Length; RW(F): Root Weight (Fresh); RW(D): Root Weight (Dry); TRL: Total Root Length; SA: Surface Area: ARD: Average Root Diameter; RV: Root Volume.
Table 5: Tassel results obtained from Mixed Linear Model (MLM).
Markers on rice chromosomes
The positions of markers on rice chromosomes were plotted in the linkage map which showing the marker location on chromosomes by using QTL cartographer. The researchers stated that the high-density linkage map is useful for the identification of quantitative trait loci responsible for various traits like yield, stress tolerance, disease and insect resistance and other traits in rice and many other crops. They have found the markers RM242 for RL and markers RM511, RM1261 and RM28130 were significantly associated with grain yield under terminal stage drought condition [18-20].
Rice is an important staple food of half of world population and because of the semi-aquatic nature of rice. Due to the change in global climatic conditions, there are many abiotic and biotic stresses in rice are arising and that is major problem in the main rice growing countries. It is an urgent need to develop the rice varieties that have an ability to withstand in drought prone areas or areas with water scarcity because drought reduces yield by 15-50% depending on the stress intensity and crop growth period at which the stress occurs in rice. The plant breeding techniques are understanding the problems and creating the drought resistance varieties. The stress can be managed by screening the drought tolerant cultivars with a focus on yield under stress in spite of the common cultivars like IR-64, IR-36, Swarna, Mahamaya and Sambha Masuri [21]. This marker also associates with many QTLs for various root morphological traits such as root thickness, maximum root length, total root weight, deep root weight, deep root weight per tiller and deep root to shoot ratio etc. [22,23]. The physical location of marker RM-408 was on chromosome-8, 125297 bp to 125487 bp which was very close to D63 dwarf gene on the short arm of chromosome-8 near the telomere [24]. The Bamleshwari genotype having higher root surface area 523 cm² which is one of the important traits in response to drought. In the earlier report data Bamleshwari found with high root length, CT9993 observed with highest total RL which was 2702.16 cm, observed in P+ 1413.6 and P- 1537.6 studies. Genotypes WR- 41, RKVY-104, Bamleshwari, Nagina-22 and CT-9993 having good root length, root volume, root diameter etc. The new techniques like association mapping or Genome-Wide Association Study (GWAS) also called as linkage disequilibrium were used widely to identify the Quantitative Trait Loci (QTL) which are responsible for drought traits. Mapping QTLs for some root architectural traits and genomic regions governing root architectural traits in non-stress and salinity stress condition facilitates the higher yield potential. Many WRKY genes play positive or negative regulatory roles in plant responses to different abiotic stresses, QTLs are identified for many important traits includes root and shoot responses, osmotic adjustment, hormonal responses, photosynthesis and drought tolerance. The SSR marker RM24894 found closely associated to the QTL qNTRG10 and qNDRW10 that facilitate marker assisted breeding [25]. In this research we have find out the markers for root traits like surface area, TRL, average root diameter, fresh root weight and dry root weight. This study is based on the rice root traits and the QTLs associated with the desirable root traits and it will also helpful in further research programs and studies such as; in drought tolerance to collect those favorable alleles into elite local line through marker assisted breeding. The software’s like STRUCTURE and TASSEL which makes the statistical studies easier and provide higher level of accuracy. The most diverse rice germplasm from their study were HUR4-3 and GSRIR1DQ125-L2-D2 and we have found that the population under study was consist of three groups viz., wild type, cultivated type and germplasm of rice. HvSSR01-80 marker, on chromosome 1 show significant association with two traits Shoot Length (SL) and Root Pulling Strength (RPS) having p-value 0.002, 0.036. The in-silico analysis revealed that this marker was present on putative QTL AQHJ009 and CQH3, the QTL responsible for root branching and penetrate root thickness [26]. They observed the RM-236 linked with rice bran content and RM-236 also associated with the aroma content, panicle number and clum length produced monomorphic allelic patterns in rice germplasm. The validation of gene-based marker-trait association for grain dimension traits RM- 505 were found polymorphic between the parents Sonasal and Pusa Basmati 1121 and linked to qgrl7.1 shows the 5.4% of phenotypic variance for grain length [27-29].
In early investigation the marker RM3825 found associated with TRL, also RM431 and RM28076 associated with root volume and average root diameter. Some markers were found with association are RM28048 for plant height and RM28130 for panicle length and plant height [30]. Our study revealed that the six markers out of 157 SSR markers showing the association with some root traits. These markers are RM408 for RW(F), RW(D) and surface area. RM505 for RW(D), RM236 for root surface area, RM-1 for TRL, NOD-1 for ARD and NOD-3 responsible for surface area. The association mapping study will be useful for future research like selection of donor and other crop improvement research.
Genome-Wide Association Mapping (GWAS) help to identify markers consistently associated with grain yield and root traits and markers RM242 and RM444 on Chr 9 using diverse accessions, thus saving resources and time needed for conventional QTL mapping effort with narrow genetic variation using a large number of biparental breeding lines. They found markers PSM52 (Chr 3), RM242 (Chr 9) and RM6909 (Chr 4) were associated with grain yield and root thickness, while marker RM444 (Chr 9) was associated with grain yield and RL [31].
The present study concluded that the markers found in this study responsible for various root related traits. The markers RM408, RM505, RM236, RM1, NOD1 and NOD3 associated with root traits like root length, average root diameter, fresh root weight, root surface area, dry root weight etc. This study will be used in further research such as donor identification and selection of candidate gene for drought traits. The genotypes WR-41, RKVY-104, Bamleshwari, Nagina-22 and CT-9993 having the good root morphology, which will be helpful for drought resistance studies. From the present investigations, we concluded that the more thick, long and branched root morphology of rice genotype will be suitable under drought conditions and more resistant to drought stress.
The authors are thankful to Rice Genome Project (RGP), Plant Molecular Biology (PMB) and Biotechnology and Richharia Research Lab (BRRL), college of Agriculture, Indira Gandhi Krishi Vishwavidyalaya (IGKV), Raipur for the genotypic data and lab facilities.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Ramteke RP, Verma SK, Agrawal T, Verulkar SB (2024) Identification of Markers for Root Traits Using Association Mapping Analysis in Rice (Oryza sativa L.). J Agri Sci Food Res. 15:167.
Received: 26-Dec-2023, Manuscript No. JBFBP-23-28658; Editor assigned: 28-Dec-2023, Pre QC No. JBFBP-23-28658 (PQ); Reviewed: 12-Jan-2024, QC No. JBFBP-23-28658; Revised: 19-Jan-2024, Manuscript No. JBFBP-23-28658 (R); Published: 26-Jan-2024 , DOI: 10.35248/2469-9837.24.15.167
Copyright: © 2024 Ramteke RP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited