Journal of Depression and Anxiety
Open Access
ISSN: 2167-1044
ISSN: 2167-1044
Research Article - (2022)Volume 11, Issue 8
Background: Depression is not only the leading cause of disability worldwide. Limited studies have been conducted to explore the proteins and chemicals associated with Age at First Episode of Depression (AFED) and Postpartum Depression (PPD) from genetic perspective.
Methods: The Polygenic Risk Score (PRS) of 130 brain proteins, 262 plasma proteins and 404 Cerebro Spinal Fluid (CSF) proteins were first calculated utilizing the genotype data from the UK Biobank. Pearson correlation analysis was then conducted to test the association of each protein PRS with AFED and PPD in UK Biobank. Based on chemical-gene interaction data of The Comparative Toxicogenomics Database (CTD), we also conducted enrichment analysis to identify the chemicals associated with depression-related proteins.
Results: Pearson correlation analysis identified 40 candidate proteins associated with AFED, such as IGFBP-7 (P=3.17 × 10-2 , Brain Protein), MIC-1 (P=1.25 × 103 Plasma Protein), 18 candidate proteins associated with PPD, such as TNFSF15 (P=6.44 × 10-4, CSF Protein), IL1R1 (P=1.17 × 10-2, CSF Protein), ART (P=1.2310-2, CSF Protein). Among 10,103 analyzed chemicals, enrichment analysis identified 43 chemicals for AFED and 13 chemicals for PPD, such as Pirinixic acid (P=7.50 × 10-4, Brian Protein) for AFED, Benzo (e) pyrene (P=7.1 × 10-3, Brain Protein) for PPD.
Conclusion: Our study identified multiple candidate proteins and chemicals associated with depression traits, which provides new clues for exploring the pathogenesis, and may serve as new targets for the treatment of AFED and PPD.
Depression; Tissue protein; Genome-wide association study; Chemicals; Comparative toxicogenomics database; Enrichment analysis
Depression is a common mental disorder whose core syndromes are depressed mood and loss of interest or pleasure [1]. According to the World Health Organization's latest report, the prevalence of depression is steadily increasing worldwide, with an estimated 322 million people (4.4% of the world’s population) living with depression. Furthermore, depression is a major contributor to the overall global burden of disease [2]. More dangerously, depression can lead to behaviors that increase the risk of death, such as suicide [3]. WHO’s Mental Health Action Plan 2013-2030 highlights the steps required to provide appropriate interventions for people with mental disorders including depression [4]. Age at First Episode of Depression (AFED) and Post-Partum Depression (PPD) are phenotypes that have not received much attention in depression studies. AFED is strongly associated with relapse of depression [5]. PPD is one of the most common birth complications. In lower- income and middle-income countries, the estimated prevalence of PPD ranges from 6.5 to 12.9% or even higher [6].
Although many studies have been conducted on depression, its etiology and pathogenesis remain unclear. So far, the widely accepted hypothesis of the pathogenesis of depression includes the monoamine hypothesis, Hypothalamic-Pituitary-Adrenal (HPA) axis disorder hypothesis, genetic environmental factors, neurogenesis, and increased secretion of inflammatory cytokines [7]. There are other possibilities that are Hippocampal Neurogenesis [8], increased inflammatory cytokines secretion [9]. Currently, the most studied protein is C-reactive protein. And there is also evidence that major depressive disorder is closely associated with increased C-reactive protein [10].
It's worth noting that previous studies have found that genetic and environmental factors are major risk factors for depression [11]. Environmental exposures such as air pollution, noise, toxins, and substance abuse might be triggers for depression. A prospective cohort study of middle-aged and older women showed that exposure to both O3 and PM 2.5 modestly increased the risk of depression [12]. The concentration of Di Ethyl Hexy l Phthalate (DEHP) metabolites was positively correlated with the risk of depressive symptoms in the elderly population [13]. Emotional and psychiatric symptoms were significantly associated with DEHP metabolites in an analysis of associations between DEHP exposure and depressive symptom subgroup scores in an older population [13]. In addition, since proteins are not only the endpoint of gene expression, but also the main biomarkers [14]. More researches on depression have focused on genes and transcriptomics, while few studied investigated depression related proteins. Therefore, we select proteins and related chemicals as entry points for depression in this study.
The development of Genome-Wide Association Study (GWAS) methods has made it a powerful tool for analyzing the more complex genetic structure of human diseases or traits [15]. In previous studies, many genetic susceptibility loci have been identified to be associated with a variety of complex diseases via GWAS including various cancers, chronic diseases, mental diseases [16]. Using the individual data from the UK Biobank, we calculated Polygenic Risk Scores (PRS) of proteins based on the significant Protein Quantitative Trait Loci (pQTLs), and then evaluate the relationships of each protein with AFED and PPD. Furthermore, based on The Comparative Toxicogenomics Database (CTD), we also explored the AFED and PPD related chemicals using enrichment analysis.
UK biobank cohort
UK Biobank is a large population-based prospective cohort study that recruited approximately 500,000 participants aged between 40 to 69 years from around the United Kingdom in 2006 and 2010. All participants were asked to report a range of demographic information and health status through questionnaires and interviews, and approved to use their anonymous data for any health-related research. The UK Biobank provides informed consent to all participants. The study was conducted under generic approval from the NHS National Research Ethics Service [17]. Our study was approved by UKB (Application 46478).
Depression trait of UK biobank
Age at first episode of depression (UKB data field 20433) was determined by asking "How old were you when you first felt like you had a two-week period, such as sadness, depression, or prolonged loss of interest in normal activities? (Whether you get any help)" to collect the results obtained. Participants with postpartum depression (UKB Data-Field 20445) were screened by asking "Does feeling depressed or losing interest occur within a few months of giving birth? Or did someone say you had postpartum depression?" The subjects who answered “yes” and “no” were analyzed as case and control groups.
UK biobank genotyping, imputation and quality control
There were 488,377 participants with genotype data in the UK Biobank. They were genotyped by either the Affymetrix UK BiLEVE Axiom Array or the Affymetrix UKB Axiom arrays. The SNP with the Hardy–Weinberg equilibrium test P>1.0 × 10-5, calling rate <98.5%, MAF <0.01 was removed. Imputation was carried out by IMPUTE4. Array design, genotyping and quality control procedures have been described in detail in previous studies [18].
GWAS of tissue protein dataset
GWAS summary data of protein levels in brain, CSF, and plasma were obtained from previous study [19]. Briefly, the study performed genome-wide association analyses of 14.06 million imputed autosomal common variants Minor Allele Frequency (MAF) ≥ 0.02 against protein levels in each tissue, in order to identify PQTLs within each tissue. They defined cis signals as those where the Single-Nucleotide Polymorphism (SNP) fell within 1 Mb upstream or downstream of the gene start site. Finally, 169, 230 and 75 independent proteins were identified in CSF, plasma and brain, respectively. Protein Quantitative Trait Loci (pQTLs) with actual p-values below the genome-wide significance (5 × 10-8) level were considered significant and included in the final list. Detailed sample information and quality control procedures have been mentioned in previous studies [19].
PRS analysis of tissue protein
Utilizing the identified pQTLs [19], the PRS of each protein of each individual was calculated as the sum of the risk alleles they carried, weighted by the effect size of the risk allele, according to the formula [20].
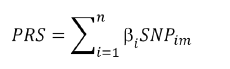
PRS denotes the PRS value of protein for UK Biobank subjects; n denotes sample size; i denotes the total number of genetic markers; βi is the effect parameter of risk allele of the significant pQTL associated with target protein, which was obtained from the GWAS of protein; SNPim is the dosage (0,1,2) of the risk allele of the ith pQTL for the mth study subject [19]. PLINK 2.0 was used to perform the PRS analysis.
Pearson correlation analysis
In the UK Biobank cohort, we used Pearson correlation analysis to evaluate associations between protein PRS and AFED or PPD. AFED were used as continuous variables, and PPD were used as categorical variables in this study. Before Pearson correlation analysis, the AFED and PPD were adjusted for sex, age, TDI, frequency of alcohol drinking per week, frequency of smoking per day and 10 Principle Components (PC) of population structure using liner regression model. All analysis was conducted using R.
Chemical-gene expression annotation dataset
The Comparative Toxicogenomics Database (CTD) (http:// ctdbase.org/downloads/) is a publicly available database designed to advance the understanding of the health effects of environmental chemicals. Based on published high-throughput experimental data and curated toxicologically important genes, CTD were established. It provides more than 1,363,085 interactions between 11,808 chemicals and 40,862 genes in 562 organisms. For this study, we downloaded the chemical-gene interaction data from CTD [21]. There were 1,788,149 annotation terms of chemical–gene pairs driven from human and mice. About 10,103 chemical-related gene sets were generated and analyzed in this study.
Enrichment analysis
By using the CTD chemical-gene expression data and the depression-related proteins, we identified the chemicals enriched in the identified proteins by using Kolmogorov-Smirnov test [22]. Normalization method control was applied to control the potential impacts of different gene sizes on enrichment analysis results [23]. The gene sets containing less than 10 genes or more than 200 genes were also excluded in this study. For the statistical test, 5,000 permutations were performed to acquire the empirical distributions of statistics for each chemical. The P-value of each chemical was calculated from the permuted empirical distribution of enrichment testing statistics [24]. Permuted P-value <0.05 was detected to be significant in this study.
Descriptive characteristics of study participants
For AFED, 81,311 participants were included; 64.56%of them were women, mean age was 55.06 years; Standard Deviation (SD) was 7.61. For PPD, 51,147 participants were analyzed, including 6,441 case samples (mean age=55.35 years, SD=7.76) and 44,706 control samples (mean age=55.08 years, SD=7.55).
Pearson correlation analysis results
For AFED, we identified 4 candidate proteins in brain, such as Insulin-like growth factor-binding protein 7 (P=3.17 × 10-2), N-acylethanolamine-hydrolyzing acid amidase (P=1.87 × 10-2); 7 candidate proteins in plasma, such as Tryptase beta-2 (P=4.90 × 10-4), Growth/differentiation factor 15 (P=1.25 × 10-3); 8 candidate proteins in CSF, such as Cathepsin B (P=1.46 × 10-2), Follicle stimulating hormone (P=2.91 × 10-2) ( Figure 1).
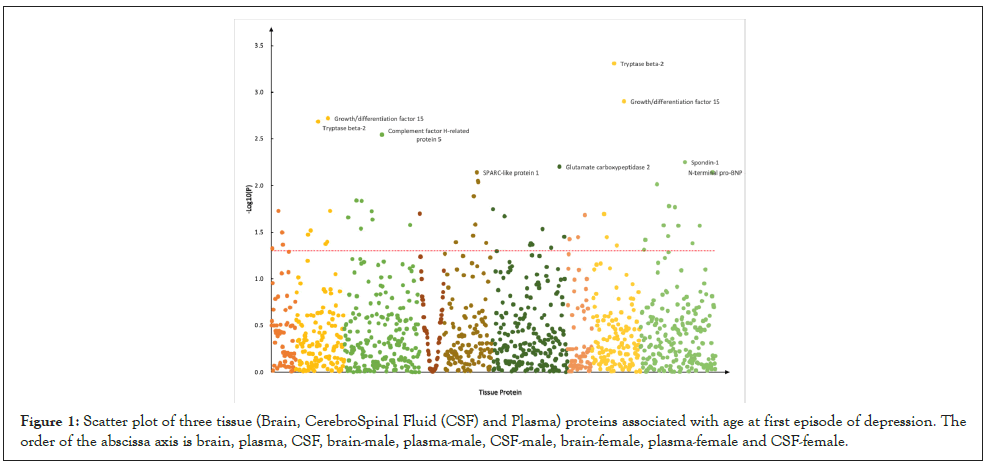
Figure 1: Scatter plot of three tissue (Brain, CerebroSpinal Fluid (CSF) and Plasma) proteins associated with age at first episode of depression. The order of the abscissa axis is brain, plasma, CSF, brain-male, plasma-male, CSF-male, brain-female, plasma-female and CSF-female.
And for PPD, we identified 1 candidate protein in brain, Leucinerich repeats and immunoglobulin-like domains protein 3 (P=2.10 × 10-2); 8 candidate proteins in plasma, such as Galectin-3 (P=1.58 × 10-2), Myeloid cell surface antigen CD33 (P=1.94 × 10-2); 12 candidate proteins in CSF, such as Tumor necrosis factor ligand superfamily member 15 (P=6.44 × 10-4), Interleukin-1 receptor type 1 (P=1.17 × 10-2) (Figure 2). Supplementary Table 1 summarizes 21 proteins that are significantly associated with PPD.
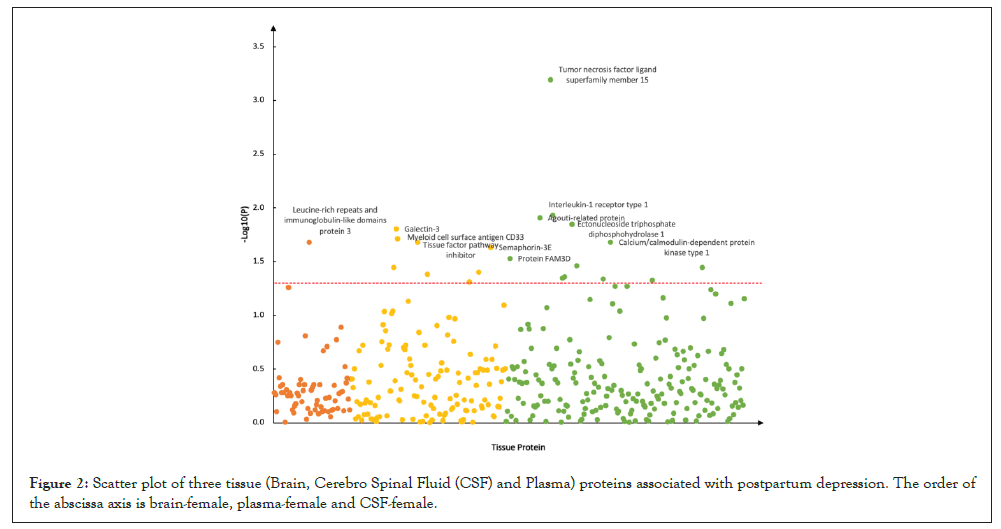
Figure 2: Scatter plot of three tissue (Brain, Cerebro Spinal Fluid (CSF) and Plasma) proteins associated with postpartum depression. The order of the abscissa axis is brain-female, plasma-female and CSF-female.
Pearson correlation analysis results by gender
In males, we found 1, 8, 9 proteins associated with AFED in brain, plasma and CSF, respectively. And in females, we found 3, 5, 12 proteins associated with AFED in brain, plasma and CSF, respectively. Table 1 shows the results of the top 10 tissue proteins associated with AFED and PPD. Supplementary Table 2 provides the detailed results of Pearson correlation analysis of AFED.
Enrichment analysis results
For AFED-related proteins, we identified 16 enriched candidate chemicals in brain, such as Pirinixic acid (P=7.50 × 10-4), Diethylhexyl Phthalate (P=2.75 × 10-3); 5 enriched candidate chemicals in plasma, such as tris (1,3-dichloro-2-propyl) phosphate (P=4.05 × 10-3), Air pollutants (P=7.70 × 10-3); 4 enriched candidate chemicals in CSF, such as Indomethacin (P=2.50 × 10-2), Belinostat (P=2.18 × 10-2) (Figure 3).
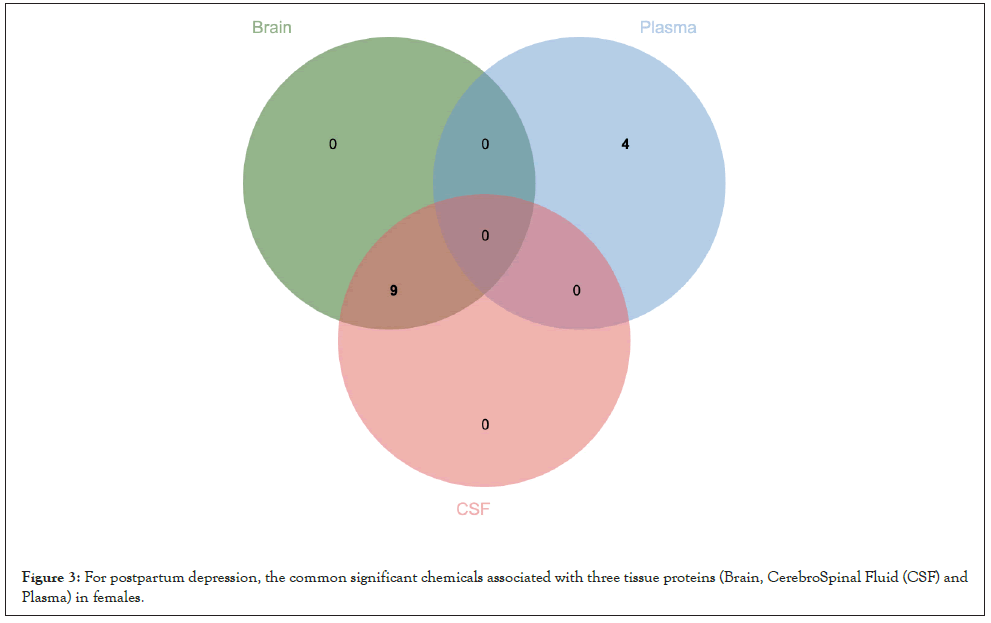
Figure 3: For postpartum depression, the common significant chemicals associated with three tissue proteins (Brain, CerebroSpinal Fluid (CSF) and Plasma) in females.
For PPD-related proteins, we identified 9 enriched candidate chemicals in brain, such as K7174 (P=6.90 × 10-3), Benzo(e)pyrene (P=7.10 × 10-3); 4 enriched candidate chemicals in plasma, such as Pirinixic acid (P=1.10 × 10-3), Phenylmercuric acetate (P=2.29 × 10-2); 9 enriched candidate chemicals in CSF, such as K7174 (P=6.15 × 10-3), Benzo(e)pyrene (P=8.45 × 10-3) (Figure 4a). And we found that Pirinixic acid (AFED: P=7.50 × 10-4, PPD: P=5.60 × 10-4) was most significantly associated with both AFED and PPD. Table 2 shows the Top 10 chemicals identified for AFED and PPD. Supplementary Table 3 summarizes 22 chemicals that are significantly associated with PPD-related proteins.
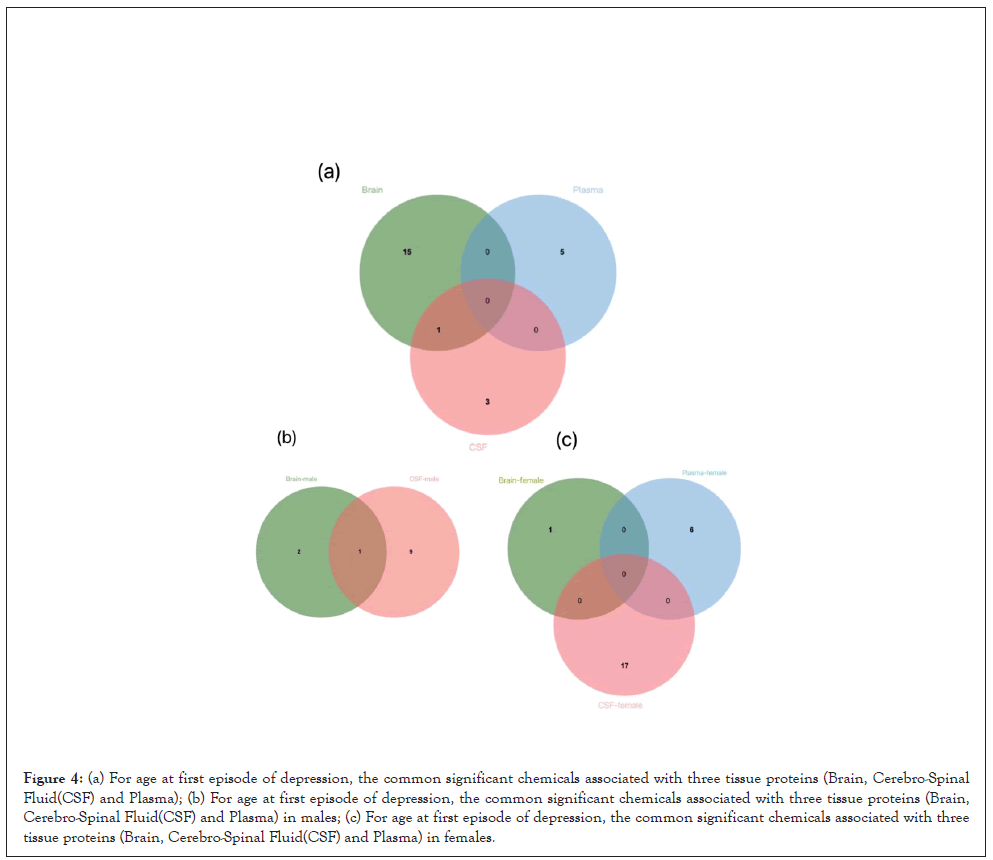
Figure 4: (a) For age at first episode of depression, the common significant chemicals associated with three tissue proteins (Brain, Cerebro-Spinal Fluid(CSF) and Plasma); (b) For age at first episode of depression, the common significant chemicals associated with three tissue proteins (Brain, Cerebro-Spinal Fluid(CSF) and Plasma) in males; (c) For age at first episode of depression, the common significant chemicals associated with three tissue proteins (Brain, Cerebro-Spinal Fluid(CSF) and Plasma) in females.
Enrichment analysis results by gender
In males, we found 3,10 chemicals enriched in AFED-related proteins in brain ,CSF, and none plasma in Figure 4b. And in females, we found 1, 6, and 17 chemicals in brain, plasma, and CSF, respectively, enriched in the AFED-related proteins (Figure 4c). Supplementary table 4 summarizes 62 chemicals that are significantly enriched in the AFED-related proteins.
In our Pearson correlation analysis, we identified multiple candidate proteins associated with AFED, such as SPARCLike protein 1(SPARCL1) and GDF15. SPARCL1 acts directly on neurons as a synaptic factor, and may act as a regeneration factor to enhance synaptic connections by increasing dendritic arborization, synapse formation and synaptic transmission [25]. In an experiment, it may in part contribute to the antidepressant-like effects of young plasma in the pathophysiology of depression [26]. Growth/Differentiation Factor 15(GDF15), also known as MIC-1, encodes a secreted ligand of the TGF-beta (Transforming Growth Factor-beta) superfamily of proteins [27] and has been implicated in various biological functions, such as metabolism [28]. In a study examining the effect of differential proteome expression on mood disorder biomarkers, the researchers found that GDF15 was higher in the bipolar depression group than in all other groups and controls [29]. And GDF15 also has been associated with cognitive impairment. In another study of depression three months after stroke, serum GDF15 levels were found to be associated with subsequent development of depression in ischemic stroke patients [27].
For PPD, the strongest associated protein is Tumor Necrosis Factor Ligand Super Family Member 15(TNFSF15) located on chromosome 9 and encodes the tumor necrosis Factor-Like Ligand 1A (TL1A) [30]. The protein encoded by this gene is a cytokine belonging to the Tumor Necrosis Factor (TNF) ligand family. The expression of this protein is inducible by TNF and IL-1 alpha [31]. TNFSF15 is best known for its association with Inflammatory Bowel Disease (IBD) [32,33]. Along this line of research, there has been interest in the relationship between a dysfunctional inflammatory response and PPD. In a recent retrospective followup analysis, TNFSF15 levels were modestly but significantly reduced in pregnant women with PPD during the first trimester of pregnancy compared to a control group without PPD [34]. Certain other members of the family have also been found to be associated with postpartum depression, such asTNFSF14,TNF,TNFSF4.
In addition, Interleukin-1 Receptor type 1(IL1R1) was also one of the most strongly protein associated with PPD. IL1R1 encodes a cytokine receptor that belongs to the interleukin-1 receptor family that provides the receptor with an endogenous innate immune system [35]. Interkeukin-1 (IL-1) plays significant roles in physiological and pathological processes in the Central Nervous System (CNS) [36], such as sleep regulation [37], memory consolidation [38] and abnormal emotional control [39]. There is increasing evidence that neuroinflammation and related immune responses are involved in the development of major depression [40, 41]. Relative to normal individuals, the mRNA expression of some specific membrane-bound receptors, such as IL1R1, TNFR1, IL1RA, was found to be significantly increased in the lymphocytes of MDD and Bipolar Disorder (BPD) patients in the depression and suicide groups [42]. Therefore, it is reasonable to speculate that IL1R1 may influence the mood change of PPD by regulating the inflammatory response involved in cytokines.
In this study, we also explored the chemicals enriched in AFED and PPD associated proteins. The top three chemicals associated with AFE associated proteins are Pirinixic acid, Potassium dichromate, and Diethylhexyl phthalate. Among them, Pirinixic acid, also known as WY-14643 , is a selective agonist of peroxisome proliferator-activated receptor-α (PPAR-α) [43]. PPAR-α that belongs to PPAR families is a ligand-activated transcription factor that could regulate neuronal survival and inflammation [44]. More and more evidence that Pirinixic acid mediates inflammatory responses are reported. Pirinixic acid can activate PPAR-α to protect cortical neurons from pro-inflammatory mediator-induced cell damage [45] and also inhibit the pro-inflammatory response of microglia [46]. In addition, in the brain, Pirinixic acid could exhibit a certain antioxidant response [47]. Based on its physiological function, a study found that Pirinixic acid pretreatment not only inhibited the production of lipopolysaccharide-induced pro-inflammatory factors, but also alleviated depression-like behaviors in mice [48].
Interestingly, the chemical most significantly associated with PPDrelated proteins was also Pirinixic acid. It means that Pirinixic acid has an important role in the development of AFED and PPD. Another very important chemical is benzo(e)pyrenes for PPD. Benzopyrene is one of the most toxic of PAHs, which are pollutants produced by the incomplete combustion of organic matter [49]. Benzopyrene easily crosses the blood-brain barrier and affects the brain's learning and memory [50]. Chronic exposure to benzopyrene can harm the nervous system and produce anxious behaviors [51]. An experiment demonstrates that benzopyrene can affect mood-related behavior in mice by affecting changes in the serotonergic and adrenergic systems [52]. Our study further confirms the association between these chemicals that affect the psychiatric system or depression and depression traits, providing new insights into risk factors for depression.
Our study may build a bridge between tissue proteins and depression, helping to provide new directions for studying the mechanism of depression at the protein level. However, it is worth noting that our study has several limitations. Firstly, all research data in our study were derived from the UK Biobank, and the research participants were limited to people of European descent. Due to different genetic backgrounds, the results of this study should be interpreted with caution when applying the results to other populations. Secondly, in our study, the sample size of significant proteins we used for Pearson correlation analysis may have been insufficient due to quality control reasons, so the results may have been compromised. Thirdly, chemical-gene interactions were queried from previously compiled literature. Thus, further large-sample prospective studies and biological studies are needed to confirm our results and elucidate the potential role of new tissue proteins in the pathogenesis of depression.
In summary, our study not only identifies multiple tissue proteins associated with both depression traits, but also multiple chemicals previously identified as being associated with the nervous system and depression. These findings provide new clues for the pathogenesis of depression and may become potential targets for the treatment and prevention of depression in the future.
Limitation of the study
The study has some limitations. First, it is cross-sectional in design, so the direction of causality between depression and associated variables could not be inferred from the findings. Second, the recall bias may occur for questions about adverse childhood experience. Thirdly, the parenting style, family dynamics, selfesteem, and negative life events of the students were not assessed. Those variables may be important factors for depression among adolescent population.
Funding information
This study was supported by the National Natural Scientific Foundation of China: 81922059 and the Natural Science Basic Research Plan in Shaanxi Province of China: 2021JCW-08.
Institutional review board statement
This study has been approved by UKB (Application 46478) and obtained participants' health-related records.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Data availability statement
The UK Biobank data are available through the UK Biobank Access Management System https://www.ukbiobank.ac.uk/. We will return the derived data fields following UK Biobank policy; in due course, they will be available through the UK Biobank Access Management System. The Comparative Toxicogenomics Database are available through the the Comparative Toxicogenomics Database Access Management System https://ctdbase.org/. We will return the derived data fields following CTD policy; in due course, they will be available through the CTD Access Management System.
Disclosure
All authors report no biomedical financial interests or potential conflicts of interest.
Author contributions
Na Zhang had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Zhen Zhang contributed equally to the work as co– senior authors.
Yan Wen, Yumeng Jia and Feng Zhang conceptualized and designed the study. All authors contributed in acquisition, analysis, and interpretation of the data. Na Zhang drafted the manuscript. Feng Zhang, Zhen Zhang helped with critical revision of the manuscript for important intellectual content. Shiqiang Cheng performed statistical analysis. Yan Wen, Yumeng Jia and Feng Zhang provided administrative, technical, or material support. Feng Zhang supervised the study.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Zhang F, Zhang N, Zhang Z, Cheng S, Jia Y, Wen Y, et al. (2022) Identifying Candidate Proteins and Chemicals for Age at First Episode of Depression and Postpartum Depression. J Dep Anxiety.11:480
Received: 05-Sep-2022, Manuscript No. JDA-22-19073; Editor assigned: 07-Sep-2022, Pre QC No. JDA-22-19073 (PQ); Reviewed: 21-Sep-2022, QC No. JDA-22-19073; Revised: 28-Sep-2022, Manuscript No. JDA-22-19073 (R); Published: 05-Oct-2022 , DOI: 10.35248/2167-1044.22.11.480
Copyright: © 2022 Zhang F, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.