
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2022)Volume 8, Issue 6
Background: Marine mollusks living in intertidal and estuarine areas, such as oysters, face numerous pathogen challenges during their development. Infection from bacteria such as Vibrio alginolyticus, represents a major factor affecting larval development and frequently leads to high mortality of the pacific oyster, Crassostrea gigas. The oyster immune response is known to play an important role in protecting the animal during development by mitigating the consequences of infection.
Results: In this study, we undertook a comprehensive analysis of the immune response of C. gigas larvae to V. alginolyticus challenge. We sequenced the transcriptome of the larval C. gigas at 0 hours which means the larval oyster without any treatment as control and 6, 12, 24, 48, and 72 hours post-infection. After RNA-seq, the raw reads are available through the NCBI Sequence Read Archive under accession number PRJNA623063. After filtering, a total of 58.24 Gb clean reads were produced and assembled using the reference genome of C. gigas. The distribution of quality Q30 was higher than 90.88% for each sample and the GC content ranged from 41.27% to 42.91%. When compared with sequences in the COG, GO, KEGG, Swiss-Prot, and NR databases, there were 1,267, 1,112, 2,187, 682, 1,133 differentially expressed genes annotated at 6, 12, 24, 48, 72 hours post-infection respectively. Numerous immune-related genes displayed differential expression that varied over time: toll-like receptors, tripartite motif proteins, Lectin-like factors, scavenger receptors, signaling pathway components such as Myeloid differentiation factor 88, and stress proteins such as Heat shock 70 kDa protein were all found to be higher in abundance following V. alginolyticus challenge compared to control. For analysis, these genes were divided into several categories such as pattern recognition receptors, fibrinogen-like proteins, damage-associated molecular patterns, complement factors, etc. We have also originally discovered many down-regulated immune-related genes in C. gigas larvae over the first 72 hours of infection, such as TRIM2, TRIM45, TRIM56, TRIM71, Macrophage mannose receptor 1, Apolipoprotein D, Cytochrome P450, Rho-related protein, Stress-induced protein 1, HSP68, HSP75, HSP70 B2, Heme-binding protein 1, Heme-binding protein 2, Heavy metal-binding protein HIP, Hemicentin-1, etc. These general categories allowed us to generate an immune response profile for C. gigas over the first 72 hours of infection. These results indicate that bacterial infection induces a complex pattern of immune gene expression in C. gigas larvae.
Conclusion: Our study will facilitate targeted investigation into the function of specific immune factors that may explain the diversity and evolution of invertebrate immune molecules and lead to the development of effective measures to improve the performance of oyster culture.
Crassostrea gigas; Bacteria V. alginolyticus challenge; Transcriptome; Immune genes
HPI: Hours Post Infection; RNA-seq: Ribonucleic Acid-sequence; COG: Groups of proteins; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; NR: Non-Redundant protein sequences; FC: Fold Change; FDR: False Discovery Rate; DEGs: Differentially Expressed Genes; MyD88: Myeloid Differentiation factor 88; Hsp70: Heat shock 70 kDa protein; TLRs: Toll-Like Receptors; PRRs: Pattern Recognition Receptors; FREPs: Fibrinogen-like Proteins; DAMPs: Damage Associated Molecular Patterns; FAO: Food and Agriculture Organization of the United Nations; TRIM: Tripartite Motif-containing; PAMPs: Pathogen-Associated Molecular Patterns; MAPK: Mitogen-Activated Protein Kinases; HSPs: Heat Shock Proteins; CR1L: Component Receptor 1-Like protein; IL17: Interleukin 17; IRAK4: IL-1 Receptor-Associated Kinase 4; TRAF 6: TNF Receptor-Associated Factor 6; NF-κB: Nuclear Factor-κB; qRT-PCR: Quantitative Real-Time PCR; EF1-α: Elongation Factor 1 α; C1q4: Complement C1q-like protein 4; SPI: Serine Protease Inhibitor dipetalogastin; AOM: Alternative Oxidase Mitochondrial; TRAFP: TNF Receptor-Associated Factor Family Protein; FC1: Fibrinogen C domain-containing protein 1; LPS: Lipopolyssacharide; PGN: Peptidoglycan; PGRPs: Peptidoglycan Recognition Proteins; SRs: Scavenger Receptors; MMR: Macrophage Mannose Receptor; LPS-BP: Hemolymph Lipopolysaccharide-Binding Protein; OsHV-1 mvar: Herpesvirus-1 microvariant; HMGB1: High-Mobility Group Box 1; MASPs: MBL-Associated Serine Proteases; TNFα: Tumor Necrosis Factor-α; IFNs: Interferons; CSFs: Colony-Stimulating Factors; CFU: Colony-Forming Units; DEPC: Diethyl Pyrocarbonate; FPKM: Fragments Per Kilobase of Transcript
The Pacific oyster Crassostrea gigas has a global distribution. Within the intertidal zone, C. gigas plays an important ecological role; however, it has also become a key commercial animal in many countries around the world [1]. In recent years, C. gigas farms in China have suffered from high and unpredictable mortality that was found to be most detrimental during the early developmental stages of the oyster. The primary cause of this mortality is exposure to the pathogenic bacteria Vibrio alginolyticus [2]. The marine, gram-negative bacterium V. alginolyticus is ubiquitous in seawater and seafood all over the world and is the main pathogen of marine culture animals such as fish, shrimp, and shellfish [3]. As V. alginolyticus is a classical standard strain in the field of marine Vibrio, however, our understanding of how oyster larvae respond to challenge by V. alginolyticus throughout developmental early stage remains scarce.
The Pacific oyster, C. gigas, is an important aquaculture oyster throughout the world, the global production of this species had expanded to 4.38 million tonnes, more than any other species of fish, mollusks or crustacea (FAO, http://www.fao.org/fishery/culturedspecies/Crassostrea_gigas/en). Because of this economic relevance, and the relative ease to culture it in controlled lab conditions, C. gigas has emerged as an important model organism for studying bivalve biology. Despite this, our understanding of the immune mechanisms that protect C.giga is still largely unknown. Based on our current knowledge, there are only a few studies that focus on advancing our understanding of the oyster immune response against bacterial challenge [4-8]. The genome of C. gigas was reported in 2012, highlighting genes involved in apoptosis, environmental stress adaptability, and shell formation. But the immune system was no given specific attention, how the immune genes changed in expression after bacterial challenge were still unelucidated [9].
Studies that have attempted to characterize the immune capabilities of C. gigas following challenge with a pathogen or Pathogen-Associated Molecular Pattern (PAMP) have primarily focused on assessing specific factors. For example, Gueguen, et al. investigated transcriptional changes in four immune-related genes of C. gigas following challenge with V. anguillarum, V. metshnikovii, V. tubiashii and V. S322 [10]. Tirape, et al. selected to focus on 18 immune-related genes from the adult C. gigas EST library and analyzed their expression pattern after Vibrio tasmaniansis, V. anguillarum and Micrococcus luteus challenge during oyster ontogenesis [6]. Song, et al. reported some immune-related genes expression change after V. splendidus infection at developmental stages of the Pacific oyster, such as phagocytosis (Integrin b-1), hematopoiesis (Gata3) , immune recognition (C-type lectin 3, TLR4 and Caspase-3) , immune elimination (Interleukin 17-5, defensin, superoxide dismutase, catalase and heat shock protein 70) and signaling transduction (REL and serine kinase I kinase) [4]. While these studies provide a few of valuable insight into the immune repertoire of C. gigas and how it is implemented in the face of infection, it does not provide a comprehensive overview of the organism-wide response.
Profiling the change of abundance of immune-related transcripts following challenge with pathogens or Pathogen-Associated Molecular Patterns (PAMPs) provides insight into how the immune response is directed against pathogen insult. Considering that bacterial infection can have serious consequences for the survival of oysters, especially during the early larvae stages [5], the information related to how the early larval stages respond to pathogen challenge remains scarce. In this study, we sequenced the transcriptome of larval C. gigas at 0 hours which means the larval oyster without any treatment as control and 6, 12, 24, 48, 72 hours post V. alginolyticus exposure by means of Illumina RNA-Seq technology. The general transcriptome was selectively analyzed to identify transcripts that displayed differential expression that varied between exposed and control animals and that also mapped to a putative immune role based on Gene Ontology (GO) searches. We found that numerous up-regulated immune-related genes displayed differential expression and many down-regulated immune-related genes in C. gigas larvae over the first 72 hours of infection that varied over time, in which, Toll-Like Receptors (TLRs), Tripartite Motif Proteins (TRIM), Lectin-like factors, scavenger receptors, signaling pathway components such as Myeloid Differentiation factor 88 (MyD88), and stress proteins such as Heat shock 70 kDa protein (Hsp70) were all found to be higher in abundance following bacterial challenge compared to control. To our knowledge, this is the first study to undertake extensively comprehensive transcriptase analyses of the immune responses of early larvae C. gigas to bacterial challenge. It will facilitate targeted investigation into the function of specific immune factors that may explain the diversity and evolution of invertebrate immune molecules and lead to the development of effective measures to improve the performance of oyster culture.
Oyster larvae and bacteria treatment
Adult individuals of oyster C. gigas were collected from a farm in Yantai, Shandong Province, China, and acclimatized in the laboratory for over one week before subsequent experiments. Sperms and eggs were scratched from gonads of male and female oysters with toothpicks. The fertilization was obtained by mixing sperms and eggs, and then the fertilized embryos were cultured in aerated seawater at 26°C.
D-hinged larvae were obtained using 300 meshes and challenged by the final concentration of 6 × 108 CFU of V. alginolyticus. Larvae without any treatment were designated as the control group. At the beginning of the bacterial challenge experiment, the samples were washed two times using filtered seawater and DEPC respectively at 1, 200 rpm. Then adding 500 μL of TRIzol Reagent (Invitrogen, USA) and stored at -80°C for later use. After bacterial challenge, the tissues were obtained by the above methods at the following time points: 6 hours post-infection (hpi),12 hpi, 24 hpi, 48hpi, and 72 hpi.
RNA extraction, cDNA library construction and illumina sequencing
Total RNA was separately isolated from the six samples using the TRIzol Reagent (Invitrogen, USA) according to the manufacturer's instructions. cDNA library construction and RNA-Seq were carried out at Beijing BioMarker Technologies (Beijing, China) based on the institute's protocols. Briefly, RNA degradation and contamination were firstly measured by 1% agarose gel electrophoresis. RNA purity was quantified by a NanoPhotometer spectrophotometer (IMPLEN, CA, USA), RNA integrity was then checked using the Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA), and RNA concentration was further determined by the Qubit RNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies, CA, USA). Based on the manufacturer's instructions, the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) were briefly described here for obtaining the sequencing libraries. Oligo (dT) magnetic beads were used to purify mRNA from total RNA and the fragmentation was randomly performed in a fragmentation buffer. Then, the first-strand cDNA was obtained by the random hexamer primer. The second strand cDNA was then synthesized by DNA polymerase I and RNase H. The end-repaired was ligated by adding the NEBNext Adaptor and dA-tailed DNA to prepare for hybridization, subsequently, the AMPure XP system (Beckman Coulter, Beverly, USA) was used to select the adaptor-ligated fragments. After PCR amplification, DNA fragments in the range of 200 to 250 bp were obtained for the following procedures. Finally, the libraries were sequenced by an Illumina HiSeq 2500 platform and 125 bp paired-end reads were obtained.
RNA sequence data analysis
For ensuring the accuracy of subsequent analysis, raw data generated from illumina sequencing were preprocessed to remove the low-quality sequence and adaptor sequences reads. After data processing, clean reads were then mapped to the reference genome (C. gigas genome) sequence using Tophat2 tools [11]. The mapped reads were assembled by Cufflinks and then the new transcripts were obtained by comparing with the annotation of the reference genome and Fragments per Kilobase of Transcript (FPKM) value was used as an indicator to measure the level of gene expression [12]. The acquired unigenes were used for a BLAST search and functional annotation of the NR, Swiss-Prot, GO, COG, KOG (Eukaryotic Orthologous Groups), KEGG, and Pfam databases.
The differentially expressed transcripts analysis
DESeq software [13] applied to experiments with biological repetition was then used to determine the differential gene expression of every 2 groups of samples with a threshold criterion of Log2 FC ≥ 1 and FDR<0.01, for up-regulated and down-regulated transcripts. The biological significance of the differentially expressed genes was determined by GO analysis. For enrichment analysis, all differentially expressed genes were mapped to terms in GO and KEGG databases, and then we searched for significantly (P ≤ 0.05) enriched GO and KEGG terms in DEGs compared with the overall transcriptome.
Validation through quantitative real-time PCR (qRT-PCR)
To confirm our illumina sequencing data, ten representative genes including Hsp70, PRR, TLR6, IL17, C1q4, MyD88, SPI, AOM, TRAFP, and FC1 were selected for qRT-PCR. Gene-specific primers of all the genes above were designed using Primer 3.0 and EF1-α gene was chosen as an internal reference gene. Quantitative RT-PCR (qRT-PCR) was carried out by QuantStudioTM 6 Flex Real-Time PCR System (Life Technologies, American) by the NovoStart® SYBR qPCR SuperMix Plus (Novoprotein Scientific Inc. China) according to the manufacturer's instructions. The data analysis was based on the Comparative threshold cycle (Ct) values of the PCR products, and the relative expression levels of the target genes were calculated as 2- Δ Δ Ct. All reactions were run in triplicate and then the statistical software SPSS 20.0 was used to analyzing the resulting Ct values by a one-way analysis of variance (ANOVA). The results were given in terms of relative mRNA expressed as means ± standard errors.
Transcriptome sequence assembly
The accession number PRJNA623063 of the raw reads are available by the National Center for Biotechnology Information Sequence Read Archive. Quality assessment of the RNA-Seq data indicated that the distribution of quality Q30 was higher than 90.88% for each sample and the GC content ranged from 41.27% to 42.91%. After filtering, 69.13% to 73.40% clean reads were successfully matched to the C. gigas genome by the TopHat2 software (Table 1). These results demonstrate that the sequencing quality was quite good, suggesting that the later transcriptome analysis results are reliable.
| Samples | Total reads | Clean bases | GC content | % ≥ Q30 | Mapped Reads | Uniq mapped reads |
|---|---|---|---|---|---|---|
| A0 | 42,488,110 | 5,351,710,454 | 41.75% | 91.46% | 31,188,582 | 19,782,843 |
| -73.40% | -46.56% | |||||
| B0 | 38,473,752 | 4,846,637,071 | 42.04% | 91.14% | 27,677,607 | 18,261,039 |
| -71.94% | -47.46% | |||||
| A6 | 40,707,324 | 5,128,064,912 | 41.90% | 91.30% | 29,169,443 | 19,418,802 |
| -71.66% | -47.70% | |||||
| B6 | 34,522,894 | 4,349,075,066 | 42.18% | 90.99% | 24,943,918 | 15,868,534 |
| -72.25% | -45.97% | |||||
| A12 | 32,554,310 | 4,100,687,702 | 41.90% | 91.10% | 23,655,300 | 14,576,967 |
| -72.66% | -44.78% | |||||
| B12 | 33,339,774 | 4,199,859,397 | 42.09% | 90.88% | 23,989,252 | 15,443,427 |
| -71.95% | -46.32% | |||||
| A24 | 33,757,068 | 4,252,398,874 | 42.91% | 91.98% | 23,601,167 | 17,022,091 |
| -69.91% | -50.43% | |||||
| B24 | 33,931,500 | 4,274,327,515 | 42.58% | 91.11% | 23,457,081 | 17,177,554 |
| -69.13% | -50.62% | |||||
| A48 | 44,693,968 | 5,630,071,580 | 41.53% | 91.23% | 32,537,843 | 18,410,125 |
| -72.80% | -41.19% | |||||
| B48 | 45,165,578 | 5,689,621,662 | 41.27% | 91.08% | 33,026,475 | 19,172,544 |
| -73.12% | -42.45% | |||||
| A72 | 44,709,090 | 5,631,977,422 | 42.29% | 91.02% | 31,729,863 | 21,427,551 |
| -70.97% | -47.93% | |||||
| B72 | 37,974,962 | 4,783,468,880 | 42.07% | 91.10% | 27,029,298 | 17,495,218 |
| -71.18% | -46.07% |
Note: A and B represent two biological repetitive samples in each group. The 0 means the larval oyster without any treatment and 6, 12, 24, 48, 72 mean the time that the oyster larvae post bacterial challenge.
Table 1: Statistical analysis of transcriptome sequencing data.
Functional annotation and classification of the Differentially Expressed Genes (DEGs) between experimental and control groups
Total, up- and down-regulated DEGs were detected between the experimental group at different time points after bacterial challenge and control. Compared with sequences in the Cluster of Orthologous Groups of proteins (COG), GO, Kyoto Encyclopedia of Genes and Genomes (KEGG), Swiss-Prot, and NCBI Non-Redundant protein sequences (NR) databases, there were 1,267, 1,112, 2,187, 682, 1,133 DEGs annotated at 6, 12, 24, 48, 72 hpi respectively (Table 2). The number of total DEGs (2,187) was the highest at 24 hpi, of which 1,745 DEGs were up-regulated and 442 were down-regulated. All the DEGs at each time point were mapped to a variety of GO annotations and three main functional classifications were determined (Figure 1): cellular component, molecular function, and biological process. Based on the GO analysis results, most immune-related genes were enriched in the category of biological process. Pathway enrichment analysis identified significantly enriched immune pathways in DEGs and showed that specific immune pathways were significantly altered following bacterial challenge (Figure 2). KEGG pathway analysis suggested that these DEGs could be assigned to specific immune-related pathways such as Toll-like receptor signaling pathway, NOD-like receptor signaling pathway, Mitogen-Activated Protein Kinases (MAPK) signaling pathway, and Apoptosis were enriched in these signaling pathways (Figures 3-7).
| DGE Set | Annotated | COG | GO | KEGG | Swiss-Prot | NR |
|---|---|---|---|---|---|---|
| 0 vs. 6 | 1,267 | 277 | 512 | 262 | 716 | 1,266 |
| 0 vs. 12 | 1,112 | 312 | 500 | 285 | 647 | 1,111 |
| 0 vs. 24 | 2,187 | 503 | 875 | 456 | 1,260 | 2,186 |
| 0 vs. 48 | 682 | 150 | 289 | 135 | 392 | 681 |
| 0 vs. 72 | 1,133 | 261 | 452 | 226 | 656 | 1,133 |
Note: The 0 means the larval oyster without any treatment and 6, 12, 24, 48, 72 mean the time that the oyster larvae post bacterial challenge.
Table 2: Table statistical analysis of differentially expressed genes.
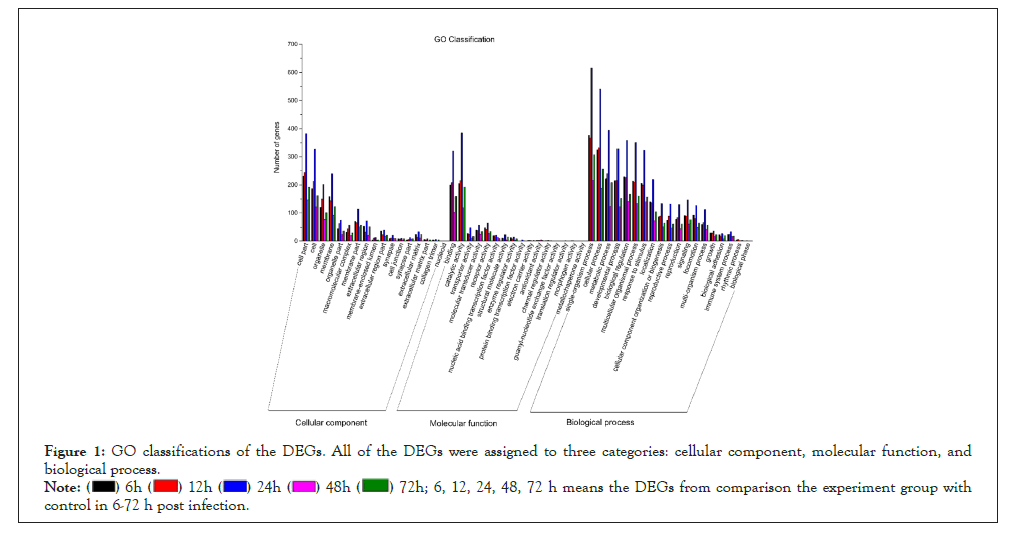
Figure 1: GO classifications of the DEGs. All of the DEGs were assigned to three categories: cellular component, molecular function, and biological process.
 6, 12, 24, 48, 72 h means the DEGs from comparison the experiment group with control in 6-72 h post infection.
6, 12, 24, 48, 72 h means the DEGs from comparison the experiment group with control in 6-72 h post infection.
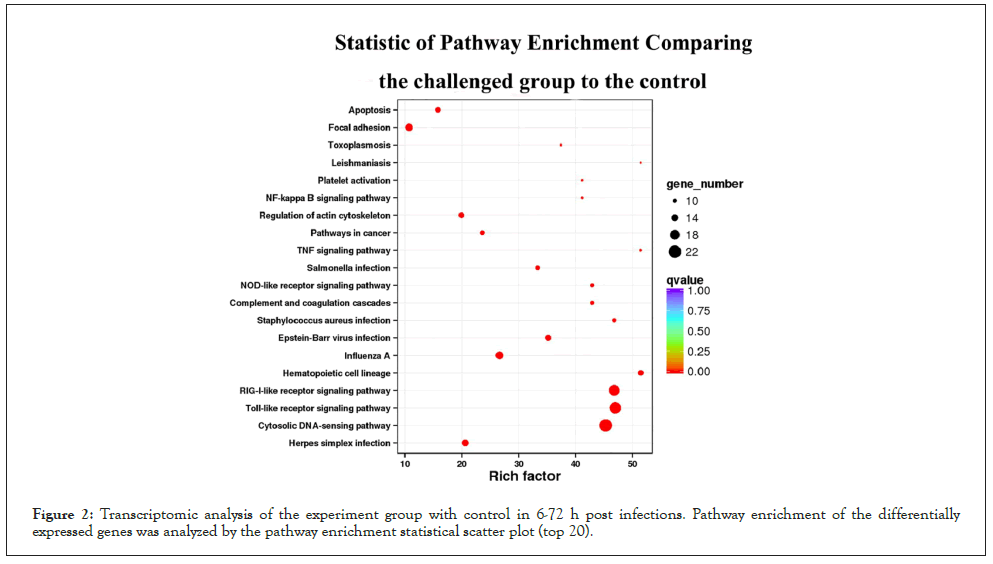
Figure 2: Transcriptomic analysis of the experiment group with control in 6-72 h post infections. Pathway enrichment of the differentially expressed genes was analyzed by the pathway enrichment statistical scatter plot (top 20).
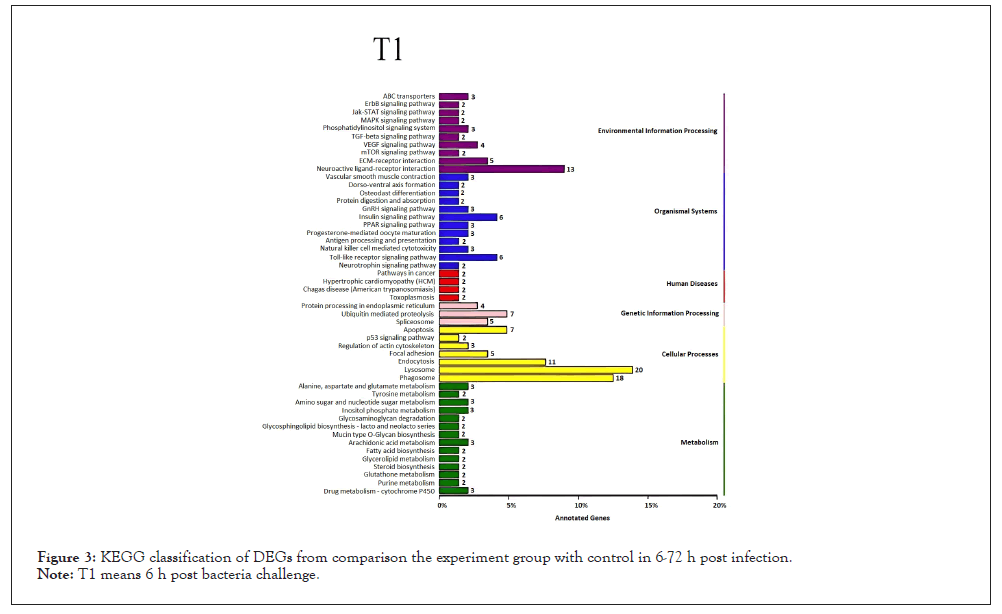
Figure 3: KEGG classification of DEGs from comparison the experiment group with control in 6-72 h post infection.
Note: T1 means 6 h post bacteria challenge.
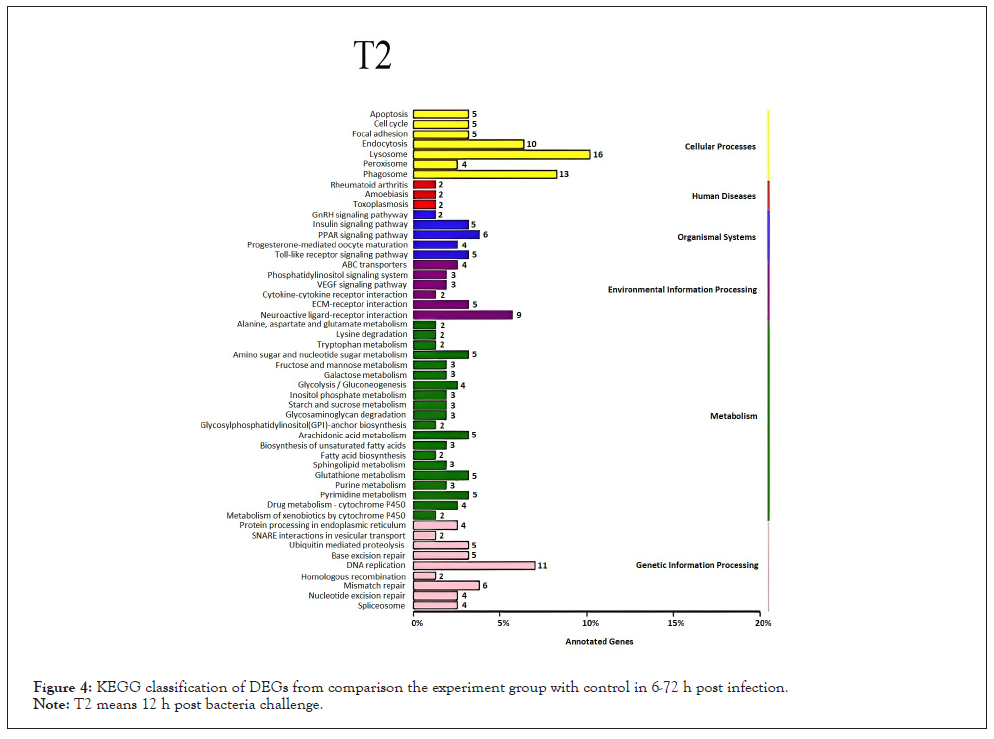
Figure 4: KEGG classification of DEGs from comparison the experiment group with control in 6-72 h post infection.
Note: T2 means 12 h post bacteria challenge.
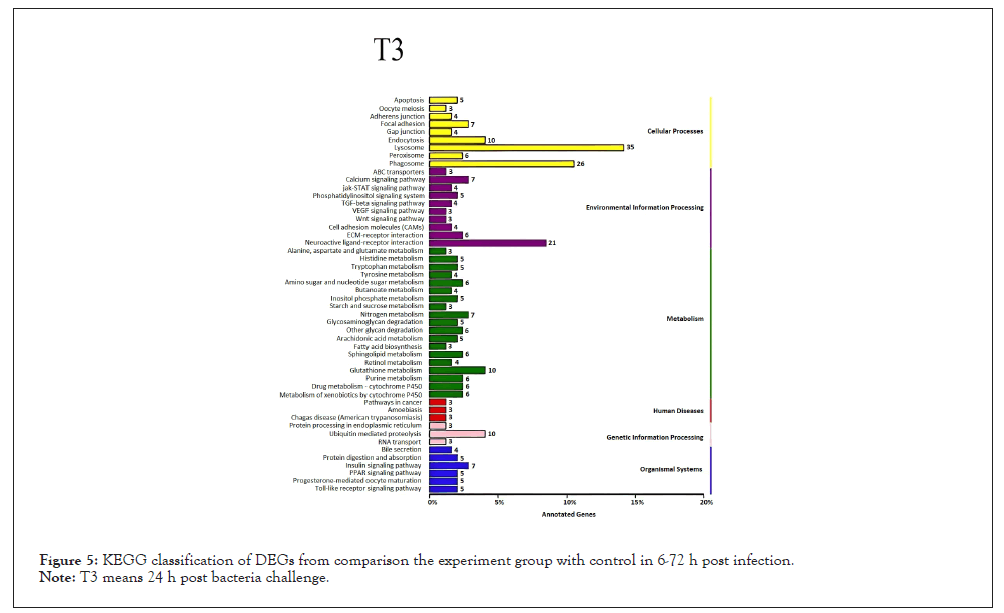
Figure 5: KEGG classification of DEGs from comparison the experiment group with control in 6-72 h post infection.
Note: T3 means 24 h post bacteria challenge.
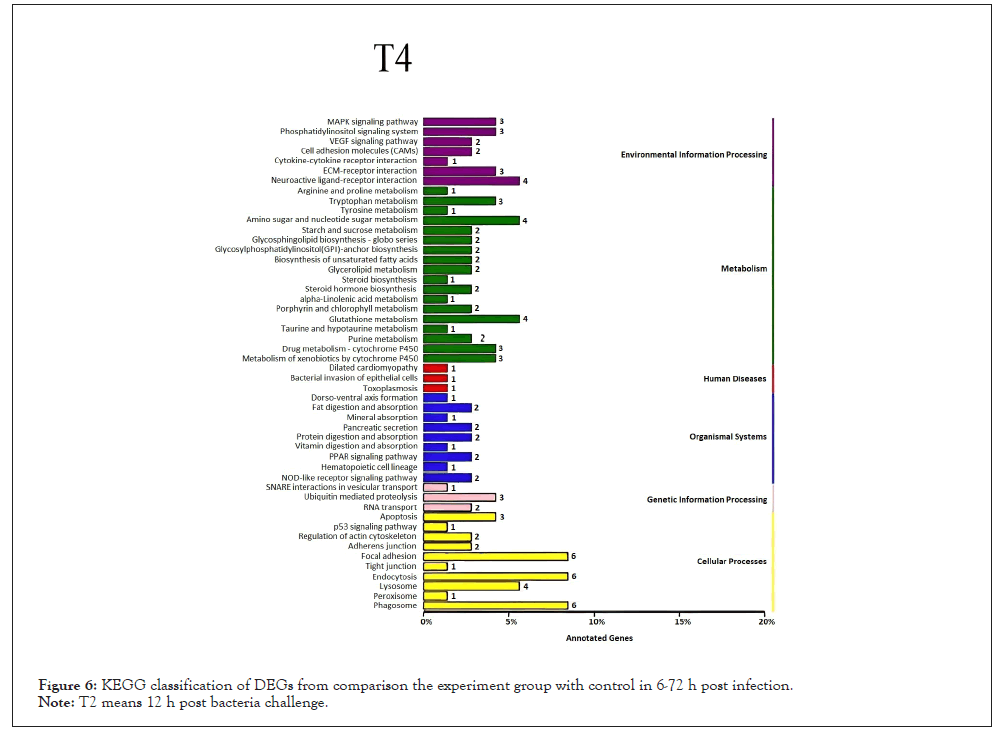
Figure 6: KEGG classification of DEGs from comparison the experiment group with control in 6-72 h post infection.
Note: T4 means 48 h post bacteria challenge.
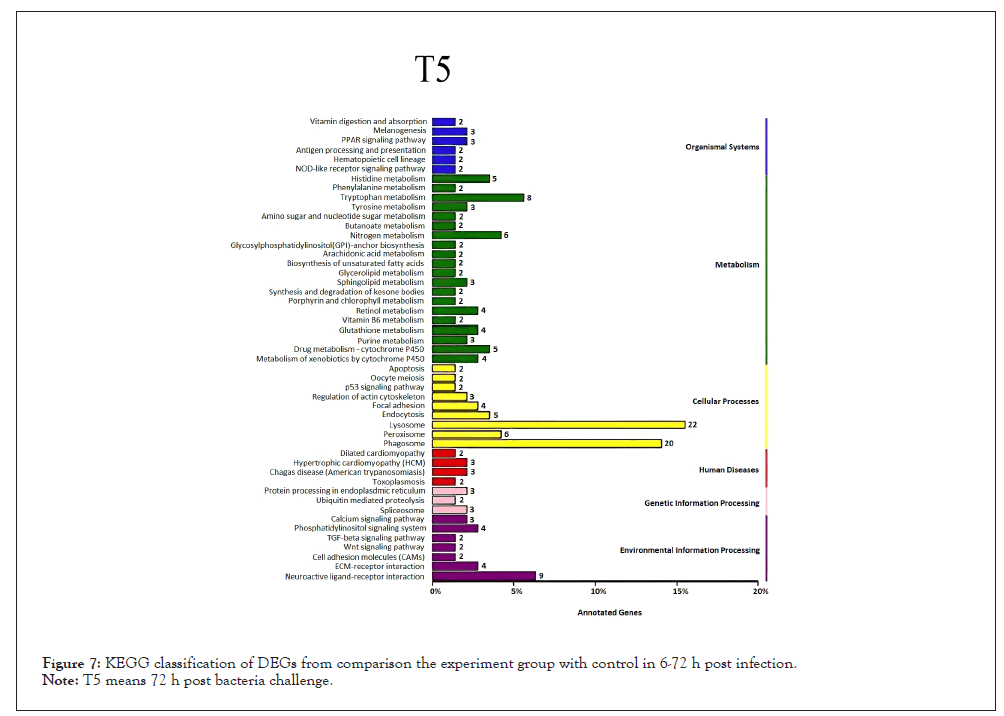
Figure 7: KEGG classification of DEGs from comparison the experiment group with control in 6-72 h post infection.
Note: T5 means 72 h post bacteria challenge.
The significantly up-regulated and down-regulated expressed immune-related genes analyses caused by V. alginolyticus
According to the analysis of GO terms and KEGG pathways, we selected the up-regulated and down-regulated immune-related genes with a threshold criterion (Log2 Fold Change (FC) ≥ 1 and FDR<0.01). Our analyses indicate that 22 immune-related transcripts were increased and 3 immune-related transcripts were down-regulated in abundance at each time point after V. alginolyticus challenge (Figures 8 and 9). Various well-known immune-related genes such as nine TLRs genes, peptidoglycan-recognition protein, six heat shock protein genes, four complement C1q protein genes, three interleukin 17-like protein genes, and five myeloid differentiation primary response protein MyD88 genes, were identified separately and divided into general categories based on their function during an immune response. These categories included; TLRs, PRRs, DAMPs, Complement, Immune effector, Cytokines, Tripartite motif-containing proteins and Immune signaling pathway.
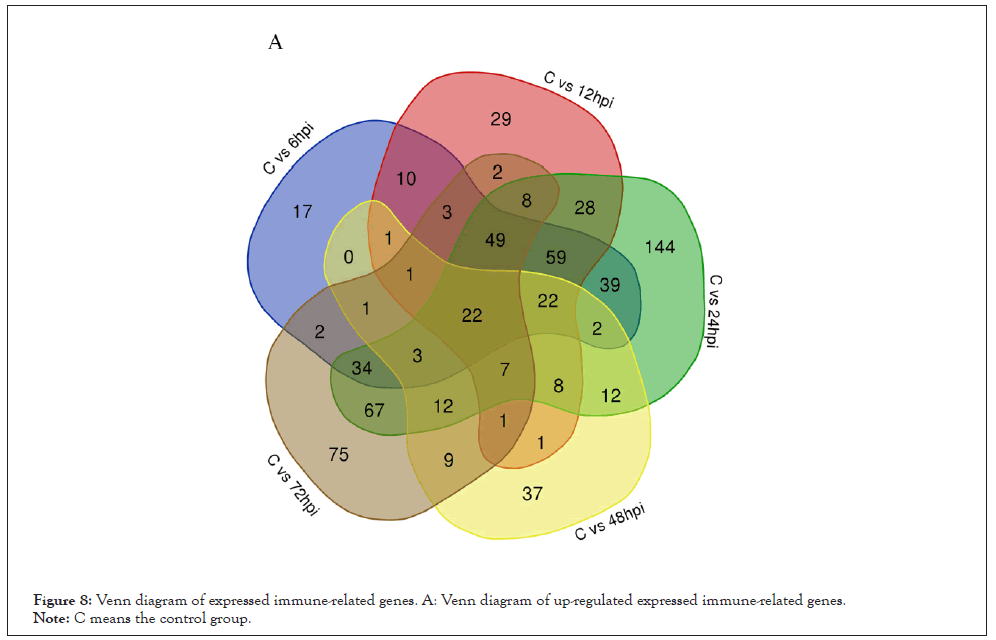
Figure 8: Venn diagram of expressed immune-related genes. A: Venn diagram of up-regulated expressed immune-related genes.
Note: C means the control group.
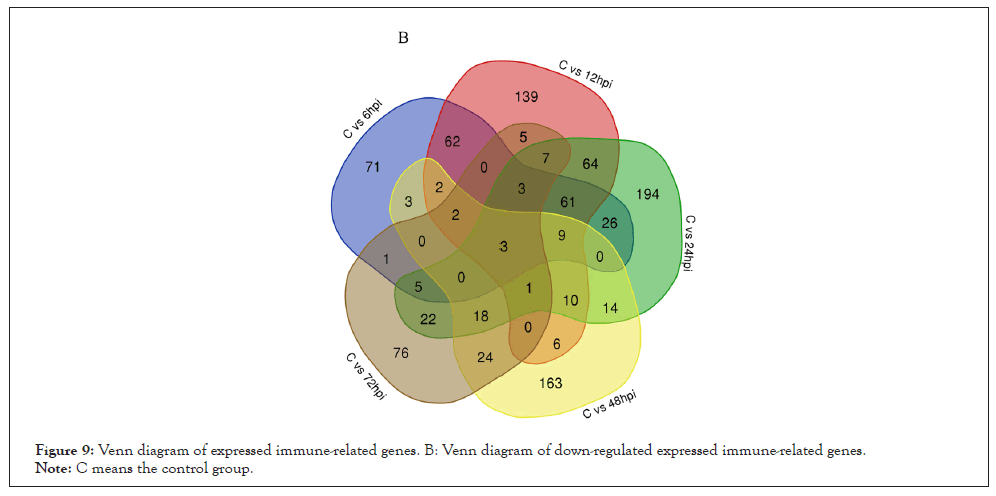
Figure 9: Venn diagram of expressed immune-related genes. B: Venn diagram of down-regulated expressed immune-related genes.
Note: C means the control group.
Pattern recognition receptors: PRRs can directly recognize and bind conserved PAMPs on the surface of invaders and play an important role in the innate immune defense response. Numerous PRRs were found in higher abundance in C. gigas transcriptome after bacterial challenge compared to controls. At nearly every time point post-challenge various PRRs were up-regulated. A majority of PRRs genes exhibited higher expression levels from 6 hpi to 24 hpi and the expression of them reduced rapidly at 48 h post-stimulation, some of them then were increased at 72 hpi. Scavenger receptor class F member 2, scavenger receptor cysteine-rich domain superfamily protein (precursor), C-type receptor 2, and macrophage mannose receptor 1 were down-regulated.
It is well known that TLRs as pattern recognition receptors play a vital role in initiating innate immune responses against pathogens challenge. A variety of TLRs expression was obviously up-regulated after bacterial challenge. The abundance of four C. gigas TLRs genes increased steadily between 6 hpi-24 hpi, then dropped gradually at 48 hpi-72 hpi, and some TLRs genes (Protein toll, TLR1, TLR6, and TLR13) were raised again at 72 hpi.
Damage associated molecular patterns: DAMPs are endogenous danger signals that can activate immune cells and originate from damaged or necrotic tissues. A common group of proteins that are often categorized as DAMPs are the Heat Shock Proteins (HSPs), which have a vital role in protecting cells against environmental stresses such as heat shock, heavy metal exposure, bacterial infection or almost any sudden changes inducing protein damage in the cellular environment [14]. In our study, we found that the transcriptome abundance of Hsp70 12A was notably increased by bacterial infection compared to controls at time points 6, 12, 24, 48, and 72h respectively. The expression level of Hsp70 12A steadily declined at 12 hpi and increased at 24 hpi, then decreased at 48 hpi and raised at 72 hpi. We also found that abundance of Hsp 68 decreased within 12 h against V. alginolyticus challenge, then the abundance of this transcript increased by bacterial infection compared to controls among 24 hpi-72 hpi.
Complement: The complement system plays a key role in innate immunity and is widely involved in physiological and pathological processes, such as phagocytosis, cytolysis, inflammation, solubilization of immune complexes, clearance of apoptotic cells and promotion of humoral immune responses [15-17]. In this study, four transcripts that reflect complement component receptor 1-like protein, complement C1q tumor necrosis factor-related protein 3, complement C1q-like protein 4 and complement C1q-like protein 3, were identified in the early larval C. gigas transcriptome after bacterial infection. Abundance of the complement Component Receptor 1-Like protein (CR1L) transcript increased gradually and reached a maximum level at 48 h post-stimulation, then declined progressively and down regulated straightly at 72 hpi. The abundance of the other three complements increased at 6 hpi and the decreased level appeared at 48 hpi, and then sharp raised up the maximum level at 72 hpi.
Tripartite motif-containing proteins: TRIM proteins have been implicated in multiple cellular functions including apoptosis, immune signaling, antiviral activity, cell proliferation, differentiation, and oncogenesis. TRIM proteins including 23 TRIM2, two TRIM3, TRIM75 and E3 ubiquitin-protein ligase displayed increased expression post bacterial infection in our study. Five TRIM2 were found to be most abundant at 6 hpi after which, the abundance dropped immediately at the other five time points, seven TRIM2 and TRIM75 were sharp reached the maximum level at 72 hpi, TRIM3 were raised the highest level at 12 hpi and then subsequently decreased progressively.
Cytokines: Cytokines are produced and secreted by cells in response to various stimulation and play vital roles in the host defense response to pathogens by performing myriad immune-relevant roles. In this study, we found three IL17 were notable up-regulated at different time points after bacterial challenge. The transcriptome expression of two IL17 steadily declined at 12 hpi and increased at 24 hpi, then dropped immediately at 48 hpi and raised at 72 hpi, the minimum level of IL17 appeared at 48 hpi.
Immune signaling pathway: MyD88 is a key adapter molecule in TLR signaling pathway and played an important role in transmitting upstream information and pathogen defense. Once MyD88 has associated with the receptor TIR domain, IL-1 Receptor-Associated Kinase 4 (IRAK4) is recruited to the receptor complex, subsequently interact with TNF Receptor-Associated Factor 6 (TRAF 6) to activate downstream signals, such as Nuclear Factor-κB (NF-κB) and MAPK, inducing the production of inflammatory cytokines and/or type I interferon to eliminate invading pathogens.
In this study, we found the expression of TLR, MyD88, TRAF family protein, and IRAK4 were significantly up-regulated following bacterial challenge. Five MyD88 genes expression were significantly increased at each time point post infection. Among these time points, MyD88 genes were expressed at the highest level at 6 hpi, then dropped immediately to a minimum at 48 hpi and subsequently increased progressively. Two TRAF family proteins were notable up-regulated at different time points post bacterial infection and one TRAF family protein was found to be most abundant at 6 hpi after which, the abundance declined gradually at the other five time points. The abundance of two IRAK4 transcripts reached a maximum level at 6 h post-stimulation, and then subsequently decreased progressively.
We have also originally discovered many down-regulated immune-related genes in C. gigas larvae over the first 72 hours of infection, such as TRIM2, TRIM45, TRIM56, TRIM71, Macrophage mannose receptor 1, Apolipoprotein D, Cytochrome P450, Rho-related protein, Stress-induced protein 1, HSP68, HSP75, Heat shock protein 70 B2, Heme-binding protein 1, Heme-binding protein 2, Heavy metal-binding protein HIP, Hemicentin-1, etc.
Validation of genes expression profiles by Quantitative Real-time PCR (qRT-PCR)
To validate the transcriptome expression profiles, ten representative up-regulation immune-related DEGs were selected for qRT-PCR analysis at 6 h, 12 h, 24 h, 48 h, and 72 h after treatment with the Elongation Factor 1 α (EF1-α) gene as the internal control (Figure 10). The overall trend of qRT-PCR-based expression patterns of ten selected genes Hsp70, Peptidoglycan-Recognition protein SC2 (PRR), TLR6, IL17, Complement C1q-like protein 4 (C1q4), MyD88, Serine Protease inhibitor dipetalogastin (SPI), Alternative Oxidase Mitochondrial (AOM), TNF Receptor-Associated Factor Family Protein (TRAFP) and Fibrinogen C domain-containing protein 1(FC1) were similar to those obtained by transcriptome analyses, indicating that the reliability of the expression profiling determined by RNA-seq was reliable and accurate, although the expression fold changes of most genes measured by qRT-PCR were more than those from transcriptome analyses.
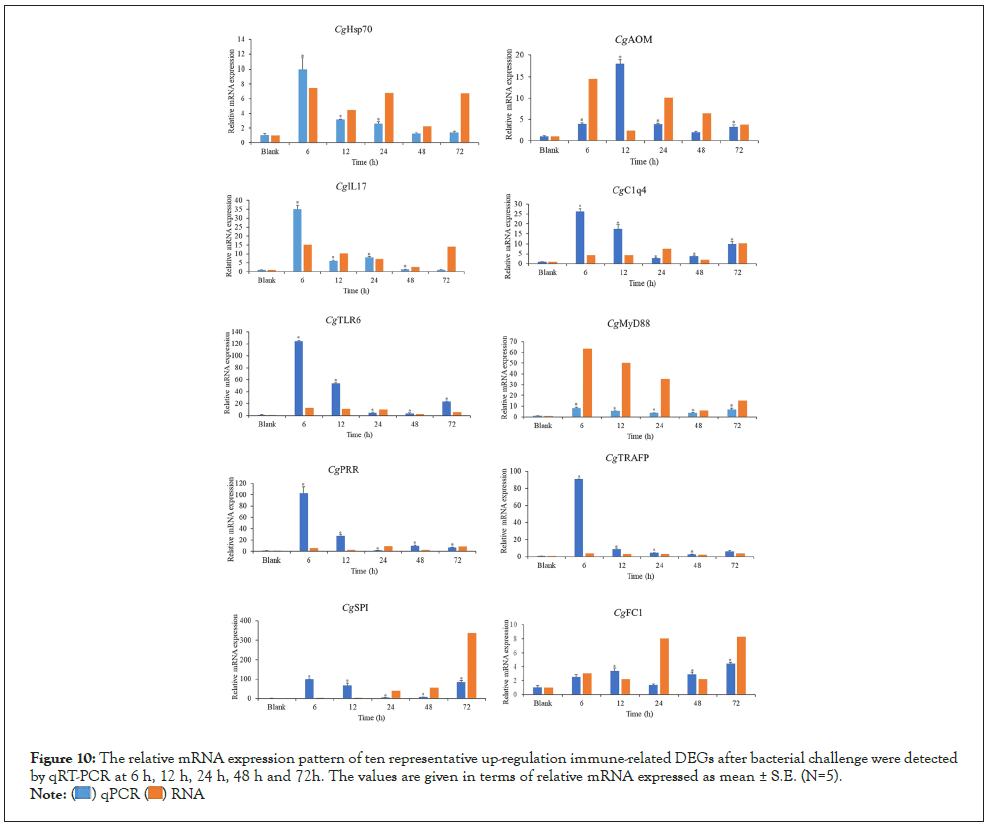
Figure 10: The relative mRNA expression pattern of ten representative up-regulation immune-related DEGs after bacterial challenge were detected by qRT-PCR at 6 h, 12 h, 24 h, 48 h and 72h. The values are given in terms of relative mRNA expressed as mean ± S.E. (N=5).
 .
.
It is well established that marine mollusks have an effective and robust immune response that protects them against infection [18]. To date, however, most of the studies that have characterized the immune responses of these animals have focused on adult individuals, while the immune response in juveniles/larvae has received less attention. For oysters and other bivalves, the developmental stages from trochophore to larvae are indispensable as calcified shells will be formed and the immune system becomes mature during this developmental stage [19]. During the early developmental periods, there can be lethal consequences because of any infectious agent may impact the developing organism. Before assessing any functional differences in the immunological capabilities between larval and adult organisms, it is important to establish a baseline of those immune factors that may be relevant during the larval stage. In present study, the expression of immune-related genes in the early larvae of C. gigas after a challenge by the bacterial pathogen V. alginolyticus was first studied to demonstrate the immune-specific transcriptome during ontogenesis.
Pattern recognition receptors
PRRs are an important part of the innate immune response. This general classification of immune receptors is defined by their ability to directly recognize different PAMPs such as Lipopolyssacharide (LPS) and Peptidoglycan (PGN) from bacteria, β-1,3-glucan from fungi, or double-stranded RNA from viruses [20]. It is the first study to identify numerous PPRs as presenting increased abundance in early C. gigas larvae transcriptome following bacterial challenge. Totally about 28 immune-related genes, such as TLRs, Peptidoglycan Recognition Proteins (PGRPs), Scavenger Receptors (SRs), Fibrinogen-like Proteins (FREPs), Calcium-dependent (C-type) mannose receptor, Macrophage Mannose Receptor (MMR), lectins, C-type receptor, protein toll, collectin-12, and hemolymph Lipopolysaccharide-Binding Protein (LPS-BP) were found in higher abundance in challenged groups compared to controls. Besides TLRs, which are discussed in detail below, PGRPs which can recognize peptidoglycan and peptidoglycan-containing bacteria play an indispensable role in innate immunity for invertebrates and vertebrates, due to its outstanding ability in detecting and eliminating invasive bacteria. It has originally found the transcription expression of PGRPs was obviously up-regulated after 24h post V. alginolyticus infection and a clearly time-dependent expression pattern of PGRPs was observed during the early development of the oyster. It is conceivable that the C. gigas PGRPs found in this study could serve not only to recognize bacteria, but also play a role in eliminating the pathogen.
Adema, et al. and Li, et al. found that a role for invertebrate FREPs in recognition of parasite-derived molecules and FREPs are proteins with at least a Fibrinogen-like (FBG) domain, they are widespread in Mollusca [21-23], and play curial roles in the innate immune response as PRRs. FREPs have been reported in the adult C. gigas [24]. We have observed that the gene expression of FREPs (Fibrinogen-like protein A and Fibrinogen C Domain-containing Protein 1) in larval C. gigas were up-regulated significantly over the first 72 hours bacterial challenge. It is inferred that the FREPs found in this study can play important roles as PRRs in the innate immune response and inhibit infection from pathogens.
SRs are a group of heterogeneous molecules on the surface of phagocytes and play an important role in innate immunity, which can recognize and mediate engulfment of a variety of pathogenic substances to eliminate the invading pathogens. Interestingly, it is a novel study to identify scavenger receptor class F member 2, scavenger receptor cysteine-rich domain superfamily protein (precursor), C-type receptor 2, and macrophage mannose receptor 1 were here down-regulated over the first 72 hours of infection in early larvae of Pacific oyster C. gigas. The down-regulation mechanism for these immune-related genes should make further efforts to study.
C-type lectin superfamily have been reported to play a potential role in the activation of complement system in the adult C. gigas [25]. C-type lectins, collectins, and macrophage mannose receptors formed this family and it was investigated that they were also associated with the cell membrane and phagocytosis [26]. Here, it was discovered that three lectin genes were up-regulated significantly post bacterial challenge during the development of the larval C. gigas. It is noteworthy that Lectin BRA-3 (Precursor) gene was up-regulated and reached a maximum level at 24 h post V. alginolyticus infection, then down regulated straightly at 48 hpi. It was inferred that the Lectin BRA-3 of the oyster larvae could be activated by V. alginolyticus to defense bacteria within 24 h post-stimulation, and then the lectin (Lectin BRA-3) was inhibited, which would suppress the oyster immune response to pathogen challenge. For oysters with the only innate immune system to defend various pathogens infection, a series of PRRs molecules might give C. gigas ability against invading pathogens.
Toll-like receptors
Tolls or TLRs play an indispensable role in initiating innate immune responses against pathogens challenge. Since the first description of Toll and its role in the immune response against fungal infection in Drosophila melanogaster [27], a large number of TLRs have been reported and functionally characterized in various species. As an ancient family of evolutionary conserved PRRs, TLRs, which are found in many species including mammals, flies, crustaceans, and mollusks, are playing as crucial roles in the immune system [28]. They play a key role in defense against invading pathogens of host early stage by recognizing conserved PAMPs and activating downstream signaling pathways which can induce the production of inflammatory cytokines and/or type I interferon to clear invading pathogens. TLR6 has been reported in adult C. gigas [29]. Here we discovered that TLRs/Tolls expression up-regulation during the development of the oyster early larvae transcriptome, including transcripts that represent protein tolls, TLR6, TLR13, TLR1, TLR2, and TLR4. Given the consistent increase in expression, it could be inferred that TLRs likely play an essential role in bacteria recognition and subsequent activation of the oyster early larvae innate immunity.
It is well established that TLRs play a considerable role in identifying invaders and their stimulation by PAMP ligands such as LPS and DAMP ligands such as Hsp70 induces a TLR signaling pathway which plays a vital role in the immune defense against pathogen infection by activating the diverse downstream reaction including anti-oxidant, anti-bacteria and apoptosis [30,31]. In the present study, we observed that the expression of TLR, MyD88, TRAF family protein, and IRAK4 were significantly up-regulated following bacterial challenge, but NF-κB has not been found in the C. gigas larvae transcriptome. MyD88 is a widespread and important adaptor for TLR/IL-1R family, and primarily recruited to activate TLR after the recognition of PAMPs/DAMPs by TLRs. Once MyD88 has associated with the receptor TIR domain, IRAK4 is recruited to the receptor complex, subsequently interacts with TRAF6 to activate downstream signals, such as NF-κB and MAPK, inducing the production of inflammatory cytokines and/or type I interferon to eliminate invading pathogens. Presently the C. gigas larvae transcriptome analyses revealed existing multiple TLR and MyD88 transcripts that demonstrate an increase in abundance following bacterial challenge, it was indicated that a MyD88-dependent TLR signaling pathway also existed in early C. gigas larvae and TLRs and the adaptor protein MyD88 respond to a range of pathogens, indicating they play an important role in the early stage innate immunity of oyster.
Damage associated molecular patterns
DAMPs are endogenous danger signals that can activate immune cells and originate from damaged or necrotic tissues. To date, various DAMPs such as HSPs and High-Mobility Group Box 1 (HMGB1) have been identified and characterized in invertebrates and vertebrates [32,33]. A common group of proteins that are often categorized as DAMPs are the HSPs. As stress proteins, HSPs are ubiquitous and evolutionary conserved, known to exist in all living organisms [34-36]. They play indispensable roles in protecting cells against environmental stresses such as heat shock, heavy metal exposure, bacterial infection, or almost any sudden changes inducing protein damage in the cellular environment [14]. According to their molecular weight, these proteins have been classified into several families, such as HSP90 (85-90 kDa), HSP70 (68-73 kDa), HSP60, HSP47, and low molecular mass HSPs (16-24 kDa) [37]. Among these proteins, HSP70s have been studied extensively and are most responsible for intracellular chaperone and extracellular immunoregulatory functions as DAMPs [33]. HSP70 has been reported in the C. gigas larvae [4]. In this study, we found HSP-70 kDa and HSP-68 kDa expression were increased during larval Pacific oyster development after bacterial challenge and a clearly time-dependent expression pattern of HSP70 was observed. The expression of the HSP70 gene increased at 6 hpi and the maximum level appeared at 24 hpi, and then dropped gradually. It is interesting that HSP68 was down-regulated within 12 h infection and then up-regulated at 24, 48, and 72 hpi. Our results suggested that HSP70 and HSP68 might be involved in oyster response to pathogenic infection and they might play an important role in the oyster immune response.
Complement
The complement system plays a key role in innate immunity against infection and is widely involved in physiological and pathological processes. The functions of the complement system include phagocytosis, cytolysis, inflammation, solubilization of immune complexes, clearance of apoptotic cells, and promotion of humoral immune responses [15-17].
There are four types of protein in the complement system, including inherent components, regulatory molecules, complement receptors, and specific protein fragments that can be activated [38]. Complement components such as C1q, mannose-binding lectin, and ficolin are PRRs that function as recognizing potential pathogens during immune responses and activating different complement pathways in vertebrates [39]. The key component of the classical complement pathway C1q involved in widespread immunological processes, such as apoptotic cell elimination, bacteria and retrovirus recognition, cell adhesion and cell growth modulation provides a major connection between innate and acquired immunity [40].
Although the complement system has been well studied in mammals, little is known about complement components in invertebrates. With the first identification of a complement homolog found in the sea urchin in 1996 [41], the recognition molecules and associated serine proteases, ficolins, MBL-Associated Serine Proteases (MASPs), C1q1, CaC1q2, C3, and C2/Complement factor B (Bf) and C1q-domain containing proteins have been recently reported in invertebrates [42-47]. In innate immunity, C1q-domain containing proteins can be considered as specialized PRPs because they can bind pathogens directly through PAMPs and to trigger phagocytosis [48]. A series of the complement related recognition molecules may be induced when foreign microorganisms invade or pathogen-related carbohydrate detection occurs in invertebrate animals. In this study, we used bacteria V. alginolyticus to infect the oyster larvae, and then analyzed the transcriptome data at different time points. With the perspective of developmental immunology, it has been found that the complement component expression was up-regulated after pathogen challenge in the oyster larvae. Interestingly, our study found that abundance of CR1L transcript increased gradually and reached a maximum level at 48 h post-stimulation, then declined progressively and down regulated straightly at 72 hpi. It was shown that the complement system of the oyster larvae could be activated by V. alginolyticus to defense bacteria within 48 h post-stimulation, then the complement system was inhibited, which would suppress the oyster immune response to pathogen challenge. Based on the result above, it could be speculated that the complement played an important role in the early stage oyster immune response.
Tripartite motif-containing proteins
More than 70 members of proteins were formed the TRIM family with three different types of domains: RING finger (R) domain, B-box (B), and a Coiled-Coil (CC) domain. The R domain of many TRIM proteins has been reported to function as E3 ubiquitin ligases by binding to both ubiquitin E2-conjugating enzymes and target proteins to facilitate the selective ubiquitination of the target, whereas the B and CC domains may be involved in protein interactions and homo/heterodimerization [49]. E3 ubiquitin ligases play as indispensable roles during ubiquitation process and are involved in the regulation of both innate and adaptive immune response [50,51].
To date, many TRIM proteins have been identified and studied well in mammals [52,53]. TRIM proteins have been involved in various cellular functions including apoptosis, immune signaling, antiviral activity, cell proliferation, differentiation, and oncogenesis. Recent studies have shown that many members of the TRIM superfamily are expressed in response to Interferons (IFNs) and are involved in a broad range of biological processes that are associated with innate immunity [54,55]. Members of TRIM family of E3 ligases are reported as important regulators of innate immunity, such as TLR signaling pathway was regulated by TRIM proteins [56,57].
However, the knowledge about TRIM proteins in mollusks is still limited. TIRM2, E3 ubiquitin-protein ligase, and ubiquitin have been identified in adult Pacific oysters [58-60]. Presently many TRIM proteins (e.g 23 TRIM2, two TRIM3, and TRIM75) and E3 ubiquitin-protein ligase as being up-regulated expression after bacterial challenge in early C. gigas larvae were identified. Five TRIM2 were found to be most abundant at 6 hpi after which, the abundance dropped immediately at the other five-time points, seven TRIM2 and TRIM75 were sharp reached the maximum level at 72 hpi, TRIM3 was raised the highest level at 12 hpi and then subsequently decreased progressively. We supposed that these TRIM proteins obtained in the present study might be involved in the oyster response to pathogenic infection by regulating TLR signaling pathway.
Cytokines
Cytokines which are produced and secreted by cells in response to various stimulation are small proteins including interleukins, IFNs, Colony-Stimulating Factors (CSFs), and TNFs. They play important roles in the host defense response to pathogens by performing myriad immune-relevant roles [61].
Few of the hallmark cytokines known from mammalian immune studies possess direct ortholog in invertebrates, however, conserved protein domains allow for invertebrate studies to target appealing candidates for functional evaluations. One cytokine that has been shown to be consistent between vertebrates and invertebrates is IL-17. Rouvier et al. first reported that IL-17 as Cytolytic T-Lymphocyte (CTL)-associated antigen 8, is a T-cell factor with proinflammatory activity [62]. It is well known that IL-17 is an important member of the proinflammatory cytokine family and plays an indispensable role in eliminating of extracellular bacteria [63]. IL17 can also activate NF-κB and MAPK signal pathways, then induce other cytokines secretion and immune cell migration, further trigger the inflammatory response [61,64]. Six IL17 genes were identified in the adult oyster [61,65]. In this study, it is the first time to report that the transcriptome expression of three novel IL17 genes dropped immediately at 48 hpi and raised at 72 hpi, the minimum level of IL17 appeared at 48 hpi during larval Pacific oyster development. It is inferred that the IL-17 found in this study can play important roles in the innate immune response and inhibit infection from pathogens.
It is very interesting here that in addition to these up-regulated immune-related genes discussed above, we have originally discovered many down-regulated immune-related genes in C. gigas larvae over the first 72 hours of infection, such as TRIM2, TRIM45, TRIM56, TRIM71, Macrophage mannose receptor 1, Apolipoprotein D, Cytochrome P450, Rho-related protein, Stress-induced protein 1, HSP68, HSP75, HSP70 B2, Heme-binding protein 1, Heme-binding protein 2, Heavy metal-binding protein HIP, Hemicentin-1, etc. Reduced abundance of transcripts (Heme-binding protein 1, Heme-binding protein 2, Heavy metal-binding protein HIP, Hemicentin-1, metal binding protein) that encode for iron binding molecules occurs in the early stage of infection (6 hpi, 12 hpi, and 24 hpi). Iron sequestration is an important resource battle between host and pathogen, and it seems as though early on in infection, the bacteria are suppressing expression of transcripts that would allow the oyster larvae to secure iron. These down-regulated immune genes might be very important for better understanding the early development stage of oyster larvae innate immunity. The down-regulation mechanism of these immune-related genes should be further study.
The transcriptome of C. gigas larvae at 0, 6, 12, 24, 48, 72 hours post V. alginolyticus exposure was sequenced through Illumina RNA-Seq technology. This is the first study to undertake comprehensive transcriptome analyses of the immune responses of C. gigas larvae to bacterial challenge. We have found that many of novel immune-related molecules such as MyD88, IL17, Fibrinogen-like protein A, Hsp70, Tripartite motif-containing protein 2, Tripartite motif-containing protein 3, TLR2, TLR4, and TLR6 were expressed very high after 6 and 12 hpi at the early stage of oyster development. Numerous immune-related genes displayed differential expression that varied over time: toll-like receptors, tripartite motif proteins, Lectin-like factors, scavenger receptors, signaling pathway components such as MyD88, and stress proteins such as Hsp70 were all found to be higher in abundance following V. alginolyticus challenge compared to control. These genes were divided into several categories such as PRRs, FREPs, DAMPs, complement factors, etc. These general categories allowed us to generate an immune response profile for C. gigas early larvae over the first 72 hours of infection. Here we have interestingly discovered many down-regulated immune-related genes in C. gigas larvae over the first 72 hours of infection, such as TRIM2, TRIM45, TRIM56, TRIM71, Macrophage mannose receptor 1, Apolipoprotein D, Cytochrome P450, Rho-related protein, Stress-induced protein 1, HSP68, HSP75, HSP70 B2, Heme-binding protein 1, Heme-binding protein 2, Heavy metal-binding protein HIP, Hemicentin-1, etc. These results will facilitate targeted investigation into the function of specific immune factors that may explain the diversity and evolution of invertebrate immune molecules and lead to the development of effective measures to improve the performance of oyster culture, especially early oyster seeding cultivation.
All authors read and approved the final manuscript. J.F, PC.H and XZ.W wrote the manuscript. HY.L, HL.C, MH.Q and JR.H commented on the manuscript. J.F, FY.M, XH.C performed and supervised the experiments. J.F and XH.C assembled, annotated, and analyzed the transcriptome data. PC.H and XZ.W contributed to the conceptualization.
We gratefully acknowledge financial support of National Natural Science Foundation of China (NO. 31272682), Middle-aged and Young Teachers’ Basic Ability Promotion Project of Guangxi (2020KY10032), and thank all members of the laboratory for helping to sample the oyster.
This research was financed by National Natural Science Foundation of China (NSFC 31272682) and Middle-aged and Young Teachers’ Basic Ability Promotion Project of Guangxi (2020KY10032).
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved. All authors were contributed equally to this work.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Fang J, Cai X, Mao F, Li H, Chen H, Qian M, et al. (2022) Immune Transcripts of the Pacific Oyster: Crassostrea gigas Larvae that Change in Expression as a Result of Bacteria Challenge. Immunotherapy (Los Angel). 8:207
Received: 28-Oct-2022, Manuscript No. IMT-22-20383; Editor assigned: 31-Oct-2022, Pre QC No. IMT-22-20383 (PQ); Reviewed: 14-Nov-2022, QC No. IMT-22-20383; Revised: 21-Nov-2022, Manuscript No. IMT-22-20383 (R); Published: 28-Nov-2022 , DOI: 10.35248/2471-9552.22.08.207
Copyright: © 2022 Fang J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.