
Journal of Clinical and Cellular Immunology
Open Access
ISSN: 2155-9899

ISSN: 2155-9899
Research - (2022)Volume 13, Issue 6
Several species of Mycobacterium have been identified as having the ability to modulate immune responses, even as heat-killed preparations. Our goal was to identify mycobacteria that could potentially act in a safe and non-toxic immune-modulating effect by promoting the production of specific chemokine and cytokine responses with a potential application for impacting the microbiome. We relied on the following Mycobacterium strains: M. smegmatis, M. agri, M. phlei, M. tokaiense, M. brumae, M. aurum, and M. obuense. M. smegmatis and M. agri were the most effective in inducing immune responses in cultured Peripheral Blood Mononuclear Cells (PBMC) manifested by extracellular productions of the cytokine IL-6, as well as the chemokines IL-8, MIP-1α and MIP-1β. Correlation analyses and immune challenges to the bacterial mixtures showed that while cytokine and chemokine responses to M. smegmatis and M. agri were similar, they were distinct from responses to either B. subtilis or Phyto-Hemagglutinin (PHA) suggesting that Mycobacterium strains and B. subtilis have different effects on the immune system. Our methodology for comparing immune responses of bacterial preparations may provide a useful tool for studying immune effects of pathogenic and non-pathogenic bacteria. Distinct immune-modulatory properties of multiple species may have potential implications for immunotherapy of cancer as well as treatments of various immune-deficiency disorders.
Mycobacteria; Cytokines; Chemokines; Immune responses; Peripheral blood mononuclear cells; B. subtilis; Immunotherapy
Mycobacteria have diverse and established significant roles in modulating immune system responses [1-3]. This includes the recognition of the impact of the Bacillus-Calmette-Guerin (BCG) vaccine which is derived from M. bovis [4]. Applications of these bacteria have been utilized for immunotherapy in the treatment of multiple types of cancer [5], as well as having molecular effects on intestinal and extra-intestinal organs, and in reference to microbiome interactions and immune-mediated diseases [6].
A number of studies have also shown that the mycobacterial cell wall can stimulate the immune system [1] and it has also been documented in killing cancer cells [7-8]. Other mycobacterial preparations were shown to induce immune responses in cultured cells [3,9-14], as well as have been used to evaluate immunestimulation activities of various Mycobacterium strains [2].
Immune-modulating Mycobacterium has been especially detailed in reference to the use of M. bovis as a consequence of the utilization of the BCG vaccine [15]. Because of the documented potential side effects of BCG [16], we sought other species of Myobacterium in an effort to determine their immune-modulating effects on peripheral blood mononuclear cells whose responsibility is to produce vital chemokines and cytokines. These were the Myobacterium strains of M. smegmatis, M. agri, M. phlei, M. tokaiense, M. brumae, M. aurum, and M. obuense. For comparative purposes, we also did parallel peripheral blood mononuclear cell challenges with B. subtilis and PHA.
Bacterial cells
M. smegmatis isolates were provided by the Institute for Tuberculosis Research, College of Medicine at the University of Illinois at Chicago. Other Mycobacterium strains were acquired from the American Tissue Culture Collection, including M. agri, ATCC27406; M. phlei, ATCC11758; M. tokaiense, ATCC27282; M. smegmatis, ATCC19420; M. brumae, ATCC51384; M. aurum, ATCC23366; M. obuense, ATCC27023; as well as B. subtilis, ATCC6051. Mycobacteria were grown in a medium containing 5 g/L L-asparagine, 5 g/L Potassium dihydrogen phosphate, 1.5 g/L Citric acid, 0.5 g/L Magnesium sulfate, 20 ml/L Glycerol, 0.2 % v/v Tween 80. The pH was adjusted to 7.4 and the medium was filter sterilized. Each strain was inoculated separately into 15 ml of the medium in 50 ml bio-reaction tubes and incubated at 37°C with shaking (220 rpm) for 3 days. A medium without inoculum was also incubated along with the mycobacteria as a blank control. After 3 days, cells were collected by centrifugation at 9000 rpm and washed three times with borate-buffered saline, pH 8.0, weighed (wet weight), and resuspended in the same buffer at 100 mg/ml The cells were heat-killed by autoclaving at 121°C for 20 minutes. The samples were also submitted for mass spectrometry analysis and the results showed no media residue in the samples (not shown). The dry-freezed B. subtilis samples were prepared by washing bacterial pellets with borate-buffered saline, pH 8.0 three times, weighed and suspended in phosphate-buffered saline, pH 7.4 at 100 mg/ml (wet weight). The samples were then autoclaved as described above.
PBMC challenges
PBMC were isolated from blood collected from healthy individuals by density gradient centrifugation as described earlier [17]. Cells were cultured at 106 cells/ml in RPMI 1640 medium supplemented with 0.5% penicillin-streptomycin solution (10,000 U/ml penicillin, 10,000 µg/ml streptomycin) 0.5% L-glutamine and 5% fetal bovine serum. One ml PBMC cultures were placed in 24-well tissue culture dishes followed by the addition of heat-killed bacterial preparations at various concentrations from 50 µg/ml to 500 µg/ml as indicated below. Ten µg/ml PHA-P (Sigma-Aldrich, St. Louis, MO) was added to separate wells as a positive control. Negative control samples contained medium only and were used for determining the basal levels of the extracellular production of cytokines and chemokines. Plates were placed in a carbon dioxide water jacketed incubator and incubated for 18 hours.
Protein assays
After the overnight culture of PBMC, 0.5 ml medium was removed from each plate, centrifuged at 16,000g at +4ºC for 2 minutes and clear cell-free liquid was placed in a minus 70ºC freezer. Cytokine and chemokine concentrations in PBMC tissue culture supernatants were measured by using multiplex immunoassay using Luminex xMAP bead array technology. Four regions of antibody- conjugated beads were used for measuring human cytokine IL-6, as well as the chemokines IL-8, MIP-1α and MIP-1β.
Statistical methods
We compared cytokine and chemokine responses in immune challenges by using the Z-test. The confidence level in all tests was set at 5%. Correlations between productions of different cytokines and chemokines in immune challenges were evaluated by using the Pearson correlation test. For this test we used normalized cytokine and chemokine concentration values to account for large differences in levels of different proteins. Each protein concentration was normalized to a mean concentration value of corresponding proteins across all analyzed challenges. A correlation of cytokine and chemokine productions in two challenges was considered strong when their r value was larger than 0.7. All statistical analyses were performed in Microsoft Excel.
M. smegmatis: Stimulated extracellular productions of cytokines and chemokines in PBMC cultures
Three isolates of M. smegmatis strains collected from feline skin, abdomen, and an abscess were characterized, grown and maintained at the Institute for Tuberculosis Research, College of Medicine at the University of Illinois at Chicago. To determine immune responses to these isolates, we isolated PBMC from healthy individuals, cultured the cells in the presence of either 50 µg/ml, 100 µg/ml or 200 µg/ml of heat-killed bacteria and measured concentrations of the cytokine IL-6 as well as the chemokines IL-8, MIP-1α and MIP-1β in tissue culture supernatants. As shown in Figures 1A-1D, all three isolates stimulated secretions of these four proteins showing a significant increase in cytokine and chemokine levels in stimulated PBMC cultures compared to controls. Protein levels did not increase at higher mycobacterial cell concentrations suggesting that the immune responses were close to the saturation levels.
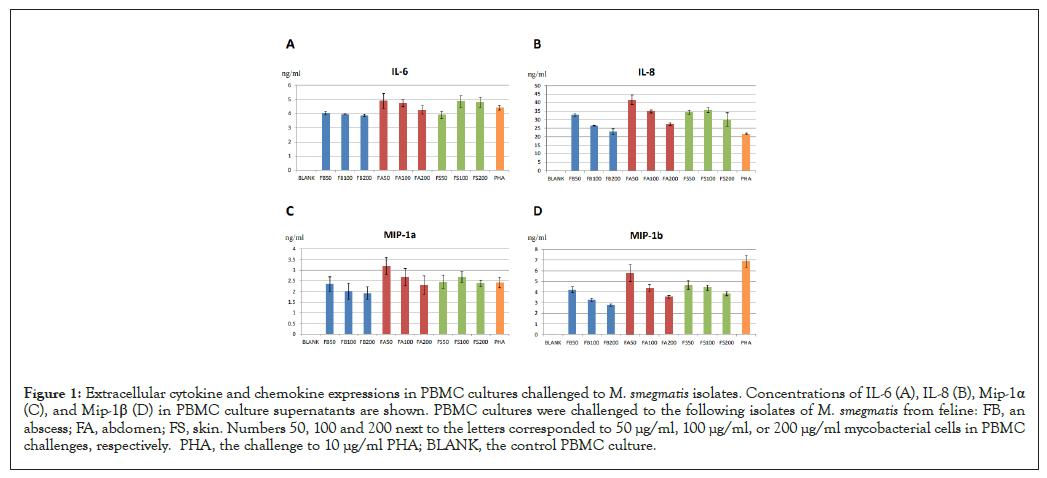
Figure 1: Extracellular cytokine and chemokine expressions in PBMC cultures challenged to M. smegmatis isolates. Concentrations of IL-6 (A), IL-8 (B), Mip-1α (C), and Mip-1β (D) in PBMC culture supernatants are shown. PBMC cultures were challenged to the following isolates of M. smegmatis from feline: FB, an abscess; FA, abdomen; FS, skin. Numbers 50, 100 and 200 next to the letters corresponded to 50 µg/ml, 100 µg/ml, or 200 µg/ml mycobacterial cells in PBMC challenges, respectively. PHA, the challenge to 10 µg/ml PHA; BLANK, the control PBMC culture.
To determine if all three isolates had the same effect on the immune cells we performed a correlation analysis of extracellular productions of different cytokines and chemokines in these challenges. The Pearson correlation test for different challenges was performed by using normalized concentrations of four proteins to correct for large differences in concentrations of different cytokines and chemokines (Figure 1A). As shown in Table 1, secretions of all cytokines and chemokines correlated well for all three M. smegmatis challenges suggesting that all three isolates evoked the same immune response. This also provided indirect evidence that these mycobacterial preparations were indeed from the same strain. In addition, we also compared Mycobacterium-specific immune responses to responses to mitogenic activator PHA [17]. The Pearson correlation test showed no or negative correlations between cytokine and chemokine levels in the PHA challenge compared to any M. smegmatis challenge suggesting that cytokine and chemokine production patterns were distinct in the PHA challenge (Table 1).
| Cell concentration | 50 µg/ml | 100 µg/ml | 200 µg/ml | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Challenge | FB | FA | FS | FB | FA | FS | FB | FA | FS |
| FB | - | - | - | - | - | - | - | ||
| FA | 0.75 | - | - | 0.77 | - | - | 0.98 | - | - |
| FS | 0.86 | 0.95 | - | 0.84 | 0.99 | - | 0.98 | 0.92 | |
| PHA | -0.51 | 0.01 | -0.03 | -0.8 | -1 | -1 | -0.6 | -0.61 | -0.6 |
Note: Numbers in bold indicate strong positive correlations.
Table 1: Spearman correlation coefficients of cytokine and chemokine expressions in PBMC challenges to M. smegmatis isolates.
M. smegmatis and M. agri challenges: Induced similar patterns of cytokine and chemokine expressions
To compare cellular immune responses to M. smegmatis with responses to other Mycobacterium strains, we challenged PBMC to either 50 µg/ml, or 100 µg/ml mycobacterial preparations from the following strains: M. agri, M. phlei, M. tokaiense, M. brumae, M. aurum, and M. obuense. M. smegmatis type strain was used as a reference. As shown in Figures 2A-2D, apart from M. smegmatis, M. agri induced high immune responses in mononuclear cells manifested by significant extracellular cytokine and chemokine productions. The concentrations of IL-6, IL-8, MIP-1α and MIP-1β were higher at 100 µg/ml of these Mycobacterium strains vs. the lower doses. Some increases in cytokine and chemokine levels were also observed in response to 50 µg/ml M. brumae. However, concentrations of these proteins decreased at 100 µg/ml M. brumae. Also, very weak responses were observed in the M. obuense challenges. We also observed significant subject-to-subject variations in cytokine and chemokine responses (compare Figure 2A to Figure 1A). The Pearson correlation test showed a strong correlation of cytokine and chemokine productions for M. smegmatis and M. agri challenges (r=0.93, Table 2). Again, no positive correlations were observed for productions of different proteins in PHA challenge compared to either mycobacteria challenge (Table 2). Similar patterns of extracellular cytokine and chemokine productions in M. smegmatis and M. agri challenges suggested that cellular immune responses to these strains were the same. In contrast, different cytokine and chemokine expression patterns in PHA challenges indicated that PHA may have induced a distinct immune response.
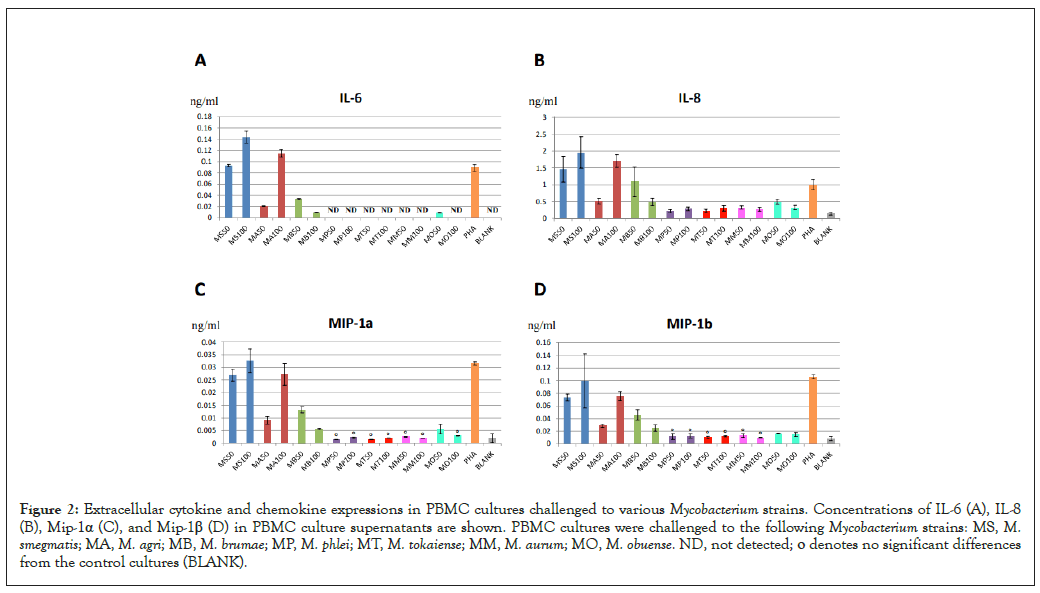
Figure 2: Extracellular cytokine and chemokine expressions in PBMC cultures challenged to various Mycobacterium strains. Concentrations of IL-6 (A), IL-8 (B), Mip-1α (C), and Mip-1β (D) in PBMC culture supernatants are shown. PBMC cultures were challenged to the following Mycobacterium strains: MS, M. smegmatis; MA, M. agri; MB, M. brumae; MP, M. phlei; MT, M. tokaiense; MM, M. aurum; MO, M. obuense. ND, not detected; ο denotes no significant differences from the control cultures (BLANK).
| Challenge | MS100 | MA100 | BS100 |
|---|---|---|---|
| MS100 | - | - | - |
| MA100 | 0.93 | - | - |
| BS100 | -0.32 | -0.29 | - |
| PHA | -0.93 | -0.95 | 0.02 |
Note: Numbers in bold indicate strong positive correlations.
Table 2: Spearman correlation coefficients of cytokine and chemokine expressions in PBMC challenges.
To further confirm that M. smegmatis and M. agri activated the same cytokine and chemokine expression profiles in immune cells, we challenged PBMC to the mixture of M. smegmatis and M. agri at 50 µg/ml each and compared cytokine and chemokine concentrations in the mixed challenge to their levels in individual challenges to 100 µg/ml bacterial cells. We hypothesized that if both Mycobacterium strains evoked the same immune response, exposures to the mixture of two strains would elevate the level of each protein to the value, which would be the average of two protein concentrations in individual challenges. Indeed, as shown in Figures 3A-3D, for all four analytes, each cytokine and chemokine concentration in PBMC cultures challenged to the mixture of two Mycobacterium strains was close to the average of their concentrations in individual challenges. The Z-test did not show any statistically significant differences between these two values for all four proteins. Our observation that the effects of M. smegmatis and M. agri challenges were not combined in the mixtures of those two strains suggested that the immune responses to these Myobacterium strains were the same.
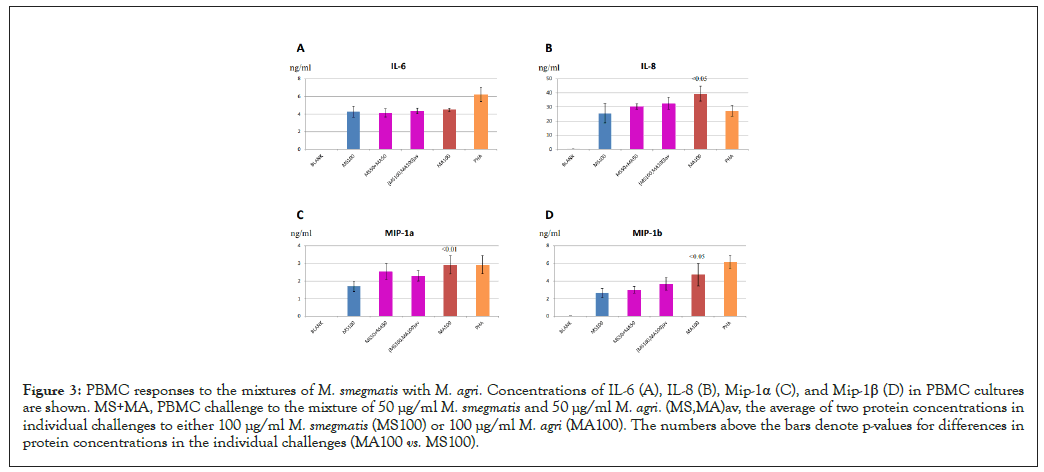
Figure 3: PBMC responses to the mixtures of M. smegmatis with M. agri. Concentrations of IL-6 (A), IL-8 (B), Mip-1α (C), and Mip-1β (D) in PBMC cultures are shown. MS+MA, PBMC challenge to the mixture of 50 µg/ml M. smegmatis and 50 µg/ml M. agri. (MS,MA)av, the average of two protein concentrations in individual challenges to either 100 µg/ml M. smegmatis (MS100) or 100 µg/ml M. agri (MA100). The numbers above the bars denote p-values for differences in protein concentrations in the individual challenges (MA100 vs. MS100).
PBMC responses to Mycobacterium strains: Distinct from responses to B. subtilis
We also compared immune responses to M. smegmatis and M. agri to responses to other bacteria of distant classification lineages. The Mycobacterium genus contains species from Actinobacteria phylum. In contrast, B. subtilis, is a Gram-positive bacteria which belongs to the distant phylum Firmicutes. Previous studies showed that B. subtilis activated immune responses in vivo [18] and induced cytokine productions in PBMC cultures [19]. To compare cellular responses to B. subtilis with responses to Mycobacterium strains, we challenged PBMC cultures to various concentrations of heat-killed B. subtilis cell preparations and measured cytokine and chemokine levels in stimulated cultures. As shown in Figures 4A-4D, B. subtilis preparations at 100 µg/ml induced productions of IL-6, IL-8, MIP-1α and MIP-1β at high levels. When cellular immune responses in B. subtilis challenges were compared with either M. smegmatis, M. agri or PHA challenges, it appeared that cytokine and chemokine productions in the B. subtilis challenge did not correlate with protein levels in other challenges (Table 2). Our results suggested that cellular responses to B. subtilis were distinct from responses to Mycobacterium strains or PHA.
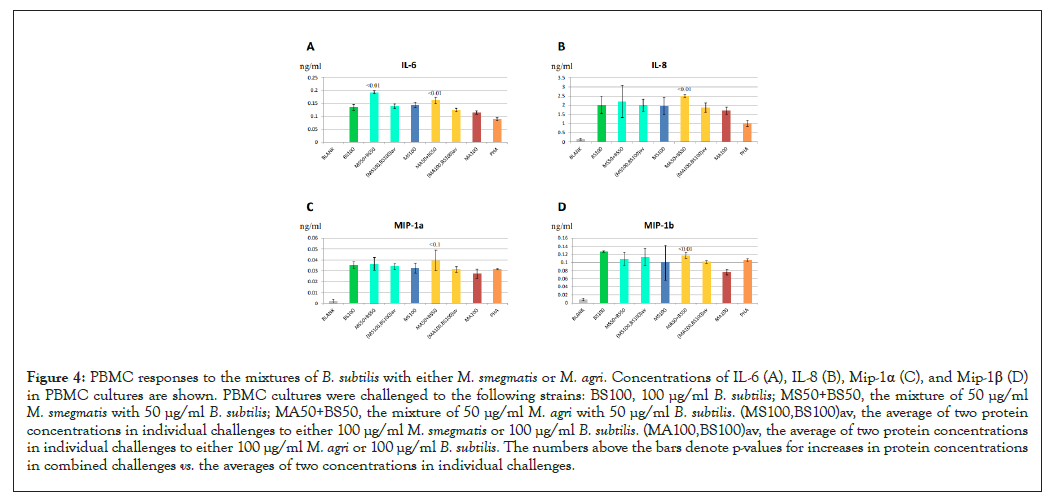
Figure 4: PBMC responses to the mixtures of B. subtilis with either M. smegmatis or M. agri. Concentrations of IL-6 (A), IL-8 (B), Mip-1α (C), and Mip-1β (D) in PBMC cultures are shown. PBMC cultures were challenged to the following strains: BS100, 100 µg/ml B. subtilis; MS50+BS50, the mixture of 50 µg/ml M. smegmatis with 50 µg/ml B. subtilis; MA50+BS50, the mixture of 50 µg/ml M. agri with 50 µg/ml B. subtilis. (MS100,BS100)av, the average of two protein concentrations in individual challenges to either 100 µg/ml M. smegmatis or 100 µg/ml B. subtilis. (MA100,BS100)av, the average of two protein concentrations in individual challenges to either 100 µg/ml M. agri or 100 µg/ml B. subtilis. The numbers above the bars denote p-values for increases in protein concentrations in combined challenges vs. the averages of two concentrations in individual challenges.
Since challenges to B. subtilis resulted in distinct patterns of cytokine and chemokine productions, this strain may have engaged a different activation mechanism of cytokine and chemokine expressions than Mycobacterium strains. In this case, a combined Mycobacterium and B. subtilis challenge would have an additive effect on cytokine and chemokine levels. To test this hypothesis we compared cytokine and chemokine concentrations in PBMC cultures, challenged to the mixture of 50 µg/ml B. subtilis and 50 µg/ml Mycobacterium preparations, with the levels of these proteins in challenges to 100 µg/ml bacterial preparations of either strain alone. Again, because higher concentrations of bacterial cells tend to suppress protein secretion (see Figure 1), the “average” effect would indicate that these two strains activate the same cytokine and chemokine production pathway, while higher than average protein levels would indicate two different activation mechanisms. As shown in Figure 4A, the cultures challenged to the mixtures of B. subtilis and either Mycobacterium strain produced significantly higher IL-6 concentrations than the averages of two IL-6 concentrations in individual challenges (Figure 4A). However, significant differences in IL-8 and MIP-1β levels were observed only in the combined B. subtilis and M. agri challenge (Figures 4B&4D). Also, differences in MIP-1α levels for mixed challenges were not significant, which may be attributed to a higher variation in measuring concentrations of this cytokine (Figure 4C). Our results demonstrated that cellular responses to B. subtilis and Mycobacterium strains were different and their combination had higher than average effects on cytokine and chemokine production. We concluded that challenges to B. subtilis strains may also engage a distinct immune response pathway, thereby resulting in partially combined effects on extracellular cytokine and chemokine productions in mixed challenges.
Weak PBMC: Responses to other Mycobacterium strains were a result of cellular toxicity or immunosuppression
We were unable to detect significant PBMC responses to M. phlei, M. tokaiense, M. aurum and M. obuense (Figures 2A-2D). We also observed decreases in cytokine and chemokine production at higher concentrations of M. smegmatis isolates (Figures 1A-1D) and M. brumae (Figure 2A). This observation raised the possibility of cellular toxicity and/or immune suppression induced by these mycobacterial preparations. To confirm or rule out this possibility we added 50 µg/ml Mycobacterium strains to 50 µg/ml B. subtilis in PBMC challenges and determined if mycobacterial cell preparations suppressed immune responses to B. subtilis. As shown in Figures 5A-5D, addition of any Mycobacterium to B. subtilis in PBMC challenges did not inhibit cytokine and chemokine productions. We also observed significant increases in protein levels in response to the M. brumae mixure, consistent with elevated responses to this Mycobacterium strain (Figures 2A-2D). Concentrations of IL-6 and MIP-1α also increased for the M. phlei mixture (Figures 5A and 5B) and a significant elevation of IL-6 levels was observed when M. tokaiense was added to the B. subtilis challenge (Figure 5A). In contrast, there was a slight statistically significant decrease in IL-8 concentrations for the M. obuense mixture (Figure 5C), consistent with the suppression of the immune response seen at the higher concentration of this strain (Figure 2A). We therefore concluded that that the lack of immune activities of Mycobacterium strains in PBMC challenges was not due to the concomitant cellular toxicity or immunosuppression induced by these bacterial preparations at concentrations tested.
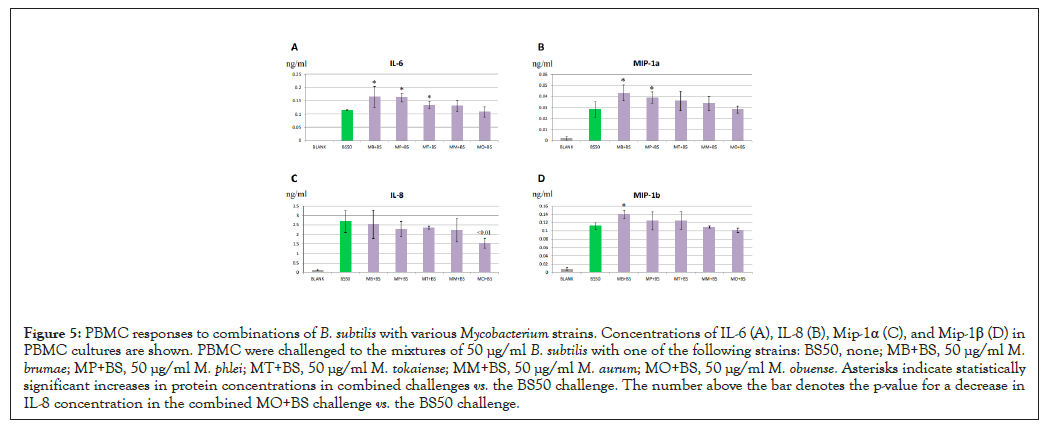
Figure 5: PBMC responses to combinations of B. subtilis with various Mycobacterium strains. Concentrations of IL-6 (A), IL-8 (B), Mip-1α (C), and Mip-1β (D) in PBMC cultures are shown. PBMC were challenged to the mixtures of 50 µg/ml B. subtilis with one of the following strains: BS50, none; MB+BS, 50 µg/ml M. brumae; MP+BS, 50 µg/ml M. phlei; MT+BS, 50 µg/ml M. tokaiense; MM+BS, 50 µg/ml M. aurum; MO+BS, 50 µg/ml M. obuense. Asterisks indicate statistically significant increases in protein concentrations in combined challenges vs. the BS50 challenge. The number above the bar denotes the p-value for a decrease in IL-8 concentration in the combined MO+BS challenge vs. the BS50 challenge.
We sought to identify potential immune-modulating properties of multiple Mycobacterium species in a heat-killed format in an effort to determine if inherent characteristics of these mycobacteria have the capacity to act in a productive immune-modulating fashion. Previous analyses of immune-modulating activities of 88 Mycobacterium strains provided useful insights regarding the utility of various strains as potential candidates for immunotherapy of cancer in reference to their pathogenicity and growth rate [2]. However, the use of the monocytic cell line for immune challenges and a limited number of immune parameters measured in this study, namely IL-12 and TNF-α, limited its applications. In the current study we evaluated the immune-stimulating properties of seven Mycobacterium strains using PBMC cultures from healthy donors and compared them to the responses to B. subtilis strain.
We measured the production of the cytokine IL-6 and, as well as the chemokines IL-8, MIP-1α and MIP-1β in immune challenges, which were the parameters previously measured in our fibromyalgia studies [17,20]. We identified two Mycobacterium strains, M. smegmatis and M. agri to be the most effective in inducing PBMC immune responses. It has been shown that M. smegmatis preparations induce a potent immune response [21], display high anti-tumor activity in a mouse model [22] and were effective in cancer immunotherapy studies [23]. Consistent with the results of our study, live M. smegmatis cells were capable of inducing the production of IL-6, IL-8 and other cytokines in neutrophil cultures [9].
M. brumae is yet another promising candidate for immunotherapy. High anti-tumor and immune-modulatory activities of this Mycobacterium strain has been demonstrated [24]. However, our results showed only moderate PBMC responses to this strain compared to M. smegmatis and M. agri (Figure 2A). The production of the cytokine IL-6 and the chemokine IL-8 by immune cells in response to M. brumae were in agreement with previously reported studies [12,25].
The absence of immune responses to M. phlei, M. tokaiense, M. aurum and M. obuense strains was somewhat surprising since these Mycobacterium strains showed significant TNF-alpha and IL-12 stimulation activities in a cultured cell line [2]. M. obuense and M. phlei were active ingredients of vaccine preparations SRL172 [26] and MCNA [27] which were investigated in a number of pre- clinical and clinical trials regarding the immunotherapy of cancer [5]. These strains were also shown to induce immune responses in human cell challenges [8,13,14,24]. The lack of PBMC responses to these mycobacteria in our experiments was not due to either cell toxicity or immunosuppression which may have been brought about by high concentrations of heat-killed cells or possible reagent contaminations. Differences in bacterial preparations or using different types of human cells may account for the apparent discrepancy of our results with previously reported studies.
We also demonstrated that cellular immune responses are similar for different Mycobacterium strains. In contrast, either PHA or B. subtilis strain of different genera evoked distinct responses in immune cell cultures, manifested by different patterns of extracellular cytokine and chemokine productions, consistent with previously reported studies [10,11]. Comparisons of cytokine and chemokine response patterns for various strains provided a useful tool in studying the immune effects of various pathogenic and non-pathogenic bacteria and determining bacterial strain identities.
The release of specific chemokines and cytokines can be especially valuable as it concerns diseases where immune deficiency exists, such as fibromyalgia, interstitial cystitis and chronic pain. If an immune-modulating intervention pathway were to be identified, various modalities of therapy with non-pathologic organisms could be achieved, thereby limiting any potential risk for adverse side effects. Sites of action can include various microbiomes including, but not limited, to the microbiome of the gastrointestinal tract and of the vagina. Mycobacterial preparations are generally safe and well tolerated [28,29]. Resultantly, the benefits of such interventions can act in a positive fashion without generating significant risks.
We identified M. smegmatis and M. agri as the most effective Mycobacterium species for inducing immune responses rendering these Mycobacterium preparations to be the most promising candidates for immunotherapy. Our results suggested that Mycobacterium strains and B. subtilis evoked distinct immune responses and have different impacts on the immune system. The distinct immune-modulating effects of Mycobacterium strains and B. subtilis may have potential implications for immunotherapy of cancer as well as the treatment of immune deficiency disorders. Our methodology for comparing immune responses for various strains may provide a useful tool for studying immune effects of various bacterial species.
We wish to thank Dr. Franzblau and Enock Mpofu from the Institute for Tuberculosis Research, College of Medicine at the University of Illinois at Chicago for providing M. smegmatis strain isolates and growing mycobacteria as well as preparing mycobacterial specimens for this study.
The authors declare no conflict of interests.
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Gavin I, Rosli F, Gillis BS (2022) Immune-Modulating Effects of Mycobacteria. J Clin Cell Immunol.13:673.
Received: 18-Nov-2022, Manuscript No. JCCI-22-20197; Editor assigned: 22-Nov-2022, Pre QC No. JCCI-22-20197 (PQ); Reviewed: 07-Dec-2022, QC No. JCCI-22-20197; Revised: 15-Dec-2022, Manuscript No. JCCI-22-20197 (R); Published: 23-Dec-2022 , DOI: 10.35248/2155-9899.22.13.673
Copyright: © 2022 Gavin I, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.