Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2022)Volume 15, Issue 4
Japanese encephalitis is transmitted by mosquitoes in humans and resultant inflammation of membranes around brain. In current study, we selected complete proteomes such as CORE PROTEIN C, PREM PROTEIN, MATRIX PROTEIN M, ENVELOPE, NS1, NS2A, NS2B, NS3, NS4B, NS4A, 2K, RNA DEPENDENT RNA POLYMERASE for T-cell epitopes identification. We found more than 100 novel T-cell epitopes for MHC class II with highest binding affinity. Out of finalized six epitopes, LRPATAWAL epitope (from Non-structural protein NS 4 B) and VRLMEAEGV epitope (from RNA-Dependent RNA polymerase) shows most negative binding energies i.e., -8.5 Kcal/mol and -8.4 Kcal/mol respectively. Results were also validated by deploying molecular dynamics and simulation studies. RMSD and RMSF values for these epitopes indicates there perfect interaction with MHC alleles. These finding provide a new insight for development of antigen based diagnostic kit and useful for crafting peptide based vaccine candidates.
Japanese encephalitis; MHC; Diagnostic; Vaccine
Japanese Encephalitis (JE) virus is a single-stranded RNA virus that belongs to the genus Flavivirus and is closely related to West Nile and Saint Louis encephalitis viruses. Japanese encephalitis is a leading cause of viral encephalitis in Asia with 30,000-50,000 clinical cases reported annually. It occurs from the islands of the Western Pacific in the east to the Pakistani border in the west, and from Korea in the north to Papua New Guinea in the south the immunity of human body is weak [1].
In a recent report suggested by Centers for Disease Control and Prevention (CDC) show that in endemic countries, where adults have acquired immunity through natural infection, JE is primarily a disease of children. However, travel-associated JE can occur among people of any age. For most travelers to Asia, the risk for JE is extremely low but varies based on destination, duration, season, and activities. From 1973 through 2011 [2], there were 58 published reports of travel-associated JE among travelers non- endemic countries. From the time of licensure of a JE vaccine in the United States in 1992 through 2011, only 7 JE cases among US travelers have been reported to CDC. Therefore, the preparation of JEV antigens is not a easy task so there are number of bioinformatics tool have been developed for the prediction of T cell epitopes in any major virulent proteins. These tools have been developed on the basis of accessible and validated data which having specific algorithms [3]. Therefore, identification of T-cell epitopes in proteins of Japanese Encephalitis (JE) virus by using Propred [4,5]. We used these tools because it is a new approach that can be used for the identification of antigenic peptide that can be further used in vaccines development. The aim of our study was to identify and map the T-cell epitopes in the complete genome proteins of JEV.
Viral proteomic selection
ViPR database [6] was deployed for screening of proteomic determinants of JEV. The sequence of all the proteins (CORE PROTEIN C, PREM PROTEIN, MATRIX PROTEIN M, ENVELOPE, NS1, NS2A, NS2B, NS3, NS4B, NS4A, 2K, RNA DEPENDENT RNA POLYMERASE) was retrieved from NCBI-GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The predictable isoelectric point (pI) and molecular weight value of proteins were also confirmed using Gene Runner and ExPasy (http://www.expasy.org/).
Epitope screening
We have used Propred [4], an immunoinformatics tool based on artificial neural networking algorithms which are being used for the identification of the T-cell epitopes from the viral proteomic domains. Such bioinformatic tool covers maximum number of human leukocyte antigen (HLA) comparison to other bioinformtaic tool. Therefore, such algorithms generate parameter during epitopes prediction such as binding efficiency threshold values ranks from 0.1 to 2.
Structural analysis and model designing
Structures of epitopes were obtained by using Pepfold 3.5 tool [7], it works on HMM (hidden markov model) algorithm and uses ab-initio approach to generate 3D model. While HLA X-ray crystallographic 3D models were retrieved from RCSB-PDB database.
Molecular docking analysis
Molecular docking was performed to further analyze the binding energies and interaction pattern between epitopes and MHC class II HLA molecules. Docking was conducted by using Dinc 2.0 website (http://dinc.kavrakilab.org/), based on AutoDock analogy [8]. This tool assisted in determining binding scores or binding energies for finalizing epitopes. Threshold for selecting epitopes interacting with HLA sets of MHC II molecules was considered below -7.0 kcal/mol [9].
Molecular dynamics and simulation
Molecular Dynamics Simulation (MDS) was conducted using the Desmond MDS suite available freely to academia [10]. The resulting complex was then solvated in TIP3P water model with orthorhombic system shape and 10 angstroms Periodic Boundary Conditions (PBC). Overall charge of the system was set to neutral through addition of appropriate Na+ and Cl- counter ions. Physiological salt concentration of 0.15 Molar was achieved through addition of NaCl. The resulting solvated system was once again energy minimized using the Desmond Minimization. The default relaxation protocol was used for preparatory phase MDS. The extent of the production MD was 100 nanoseconds and the conformations were recorded every 100 picoseconds. Root mean square deviation/fluctuation (RMSD/F) and principal component analysis were done through R based BIO3D module. Structural artworks were created through Pymol and VMD.
JEV is global health problem and suppressed the human immune system that allows more resistance of infection. In this study, all proteins of JEV were used for physicochemical analysis such as molecular weight, isoelectric point (Pi value) and antigenic nature. RNA DEPENDENT RNA POLYMERASE protein showed the highest molecular weight 103303.91 kDa while the lowest molecular weight was 2360.79 kda of 2K3 protein. Isoelectric point of these proteins was ranged between 6.10-8.89. The physicochemical properties of proteins of JEV were given (Table 1).
| Protein designation | Accesion number | Isoelectric point | Molecular weight in KDA |
|---|---|---|---|
| Core protein C | NP_775662.1 | 12.61 | 11726.24 |
| Pre M Protein | NP_775664.1 | 8.36 | 19009.79 |
| Matrix protein M | NP_775665.1 | 10.05 | 8329.63 |
| Envelope | NP_775666.1 | 7.35 | 53456.74 |
| NS 1 | NP_775667.1 | 6.44 | 40008.20 |
| NS 2 A | NP_775668.1 | 10.51 | 24540.55 |
| NS 2 B | NP_775669.1 | 4.33 | 14308.69 |
| NS 3 | NP_775670.1 | 7.14 | 68737.87 |
| NS 4 A | NP_775671.1 | 7.70 | 13687.53 |
| 2 K | NP_775672.1 | 6.10 | 2360.79 |
| NS 4 B | NP_775673.1 | 9.63 | 27073.6 |
| RNA dependent RNA Polymerase | NP_775674.1 | 8.89 | 103303.91 |
Table 1: Physicochemical properties of different putative proteins of JEV.
Epitope screening
In our study, we have used all proteins of JEV (CORE PROTEIN C, PREM PROTEIN, MATRIX PROTEIN M, ENVELOPE, NS1, NS2A, NS2B, NS3, NS4B, NS4A, 2K, RNA DEPENDENT RNA POLYMERASE) which is encoded by genome of JEV. It was used for identification of T-cell epitopes. The identification of epitopes in complete proteomic set of JEV was predicted by PROPRED for MHC class II respectively (Table S1).
Similar approach has been previously used for T-cell epitopes prediction from human papillomavirus, tick borne encephalitis virus [11], influenza A virus [12] and mycobacterium tuberculosis [13].
Molecular docking analysis
Molecular docking studies reveal binding energies between selectable epitope and HLA molecule. Best six docked complexes enlisted in Table 2. Binding pocket interaction analysis between MHC Class II molecule (HLA-DRB1*15:01) and all six selected epitopes validate significant role of these epitopes to elicit immune response. This study provide easiest and fastest approach for determining epitope based peptide vaccine candidates for developing possible therapeutic strategies against Japanese encephalitis virus (Figure 1 and Table 2).
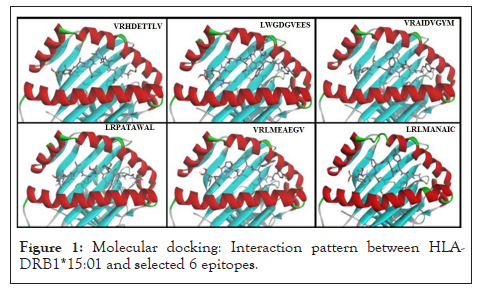
Figure 1: Molecular docking: Interaction pattern between HLA- DRB1*15:01 and selected 6 epitopes.
| Docked model | Binding energy (Kcal/mol) |
|---|---|
| VRHDETTLV-HLA DRB1*15:01 | -7.8 |
| LWGDGVEES- HLA DRB1*15:01 | -7.9 |
| VRAIDVGYM-HLA DRB1*15:01 | -8.3 |
| LRPATAWAL-HLA DRB1*15:01 | -8.5 |
| VRLMEAEGV- HLA DRB1*15:01 | -8.4 |
| LRLMANAIC - HLA DRB1*15:01 | -8.1 |
Table 2: Binding energies for best docked epitope and HLA sets of MHC class II molecules.
Molecular simulation analysis
Molecular dynamics and simulation studies provide a suitable range of RMSD and RMSF values for docked complexes, LRPATAWAL- HLA DRB1*15:01 and VRLMEAEGV-HLA DRB1*15:01. These values suggest perfect interaction between epitopes and MHC allele for long 100 ns run (Figure 2) [14].
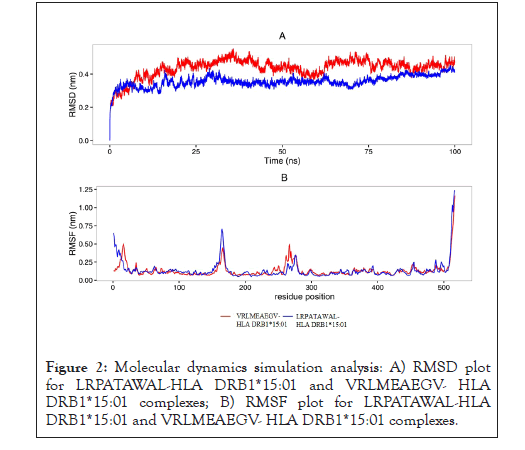
Figure 2: Molecular dynamics simulation analysis: A) RMSD plot for LRPATAWAL-HLA DRB1*15:01 and VRLMEAEGV- HLA DRB1*15:01 complexes; B) RMSF plot for LRPATAWAL-HLA DRB1*15:01 and VRLMEAEGV- HLA DRB1*15:01 complexes.
JEV is a major human health concern and an urgent need arises for development of susceptible and precise diagnostic. Thus, immunoassays are required purified antigen which is possible by using culture of JEV or recombinant technology. To avoid this problem, bioinformatics is a good approach for prediction of epitopes without using wet laboratory practice. In our study, we determined two possible epitopes LRPATAWAL and VRLMEAEGV that can perfectly interact with MHC class II alleles, and elicit immune response. Here we not only used molecular docking approaches, but also advanced simulation studies to validate our findings.These findings provide a new approach for vaccine development which minimize the labour, cost and time for diagnosis of JEV. Our approach can help to accelerate virology research for the development of vaccine. These novel T-cell epitopes can be chemically synthesized and used as antigen for JEV diagnostic and can be also used for development of peptide based vaccine.
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar].
[PubMed].
Citation: Deval R, Kumar V, Joshi A (2022) Immuno-Informatic Study: Screening of Putative Therapeutic T-Cell Epitopes against Japanese Encephalitis Virus. J Proteomics Bioinform.15:582.
Received: 13-Apr-2022, Manuscript No. JPB-22-16795; Editor assigned: 18-Apr-2022, Pre QC No. JPB-22-16795 (PQ); Reviewed: 02-May-2022, QC No. JPB-22-16795; Revised: 09-May-2022, Manuscript No. JPB-22-16795 (R); Published: 16-May-2022 , DOI: 10.35248/0974-276X.22.15.582
Copyright: © 2022 Deval R, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.