Immunome Research
Open Access
ISSN: 1745-7580
ISSN: 1745-7580
Research Article - (2020)Volume 16, Issue 2
Treponema pallidum is a gram negative bacteria and the main cause of syphilis which is classified as chronic inflammatory discompose antecedent transmitted sexually. Syphilis affects the central nervous system and the cardiovascular system, potentially leading to hearing or visual loss, aortic aneurism, stroke-like syndrome, dementia and paralysis. T. pallidum has the ability to stimulate adaptive immune and corresponding innate procedures in tissue and blood that might set the era for the HIV’s bidirectional transmission. This study expects a real epitope-based vaccine against β-Barrel outer membrane protein of Treponema pallidum designed by immunoinformatics approaches. The sequences were saved from NCBI and a number of prediction tests were undertaken to explore possible epitopes for B-cell, T-cell MHC class I and II. 3D structure of the most hopeful epitopes was illustrated. Two epitopes showed high binding affinity for B-cells, while five epitopes showed high binding affinity for MHCI and MHCII. The results were hopeful to formulate a vaccine with 71.88% world population coverage. We expect that these hopeful epitopes helps as a preventive formula for the disease in the future and recommend further in vitro and in vivo studies
Immunoinformatics; Treponema pallidum; Outer membrane beta-barrel; Peptide vaccine; Epitope
Syphilis is chronic inflammatory discompose antecedent transmitted sexually by the spirochete Treponema pallidum [1-8]. There are four subspecies for Treponema (i.e., subsp. pallidum [syphilis], subsp. endemicum [bejel], and subsp. pertenue [yaws]) and T. carateum (pinta) [9,10]. T. pallidum is vastly motile extracellular bacterium distinguished for its invasiveness, immuno-evasiveness and persistence [11]. Based on data from 2016, the WHO assessments showed that approximately 90 million people who were from China [12], Africa, Asia and the Western Pacific are currently at risk of yaws. Globally, around eleven million [2,10,11,13] rates among commercial sex workers are between 23% and 32%. In the United States [2], people were diagnosed with syphilis in 2008, with mother-tochild conduction occurring in nearly two million pregnancies [14]. Syphilis influence in statement progress to affect the cardiovascular and central nervous systems of infected individuals, potentially leading to aortic aneurism, hearing or visual loss, dementia, paralysis, and stroke-like syndrome. T. pallidum has the facility to stimulate corresponding innate and adaptive immune procedures in blood and tissue that could set the period for the bidirectional transmission of HIV [3,15]. The diagnosis of syphilis principally relies on serological testing for reactivity to treponemal and non-treponemal (cardiolipin) antigens. Indirect detection or examination of cerebrospinal fluid (CSF) of the patients can be used. However, they are often insensitive and difficult to enter [13]. Direct detection methods like dark-field microscopy, direct fluorescent antibody testing and nucleic acid amplification testing [16].
The hidden mechanism within syphilis remains ambiguous due to the main causative agent of the disease which cannot be cultured in the laboratory. So manipulation of genetic approach is important [17,18]. Gram negative bacteria are characterized by a unique membrane of integral protein that is divided into β-barrel structure consisting of eight to twenty six antiparallel amphipathic strands [19,20]. The main function of the β–barrel outer membrane (OMPs) dynamic for membrane biogenesis and transport signaling, enzyme and receptor [21,22]. β -Barrel outer membrane protein is critical target to conserved domain region and contains N-and C-terminal domains (TprCN and TprCC, respectively) [11].
Currently, the medical research has improved by immunoinformatics technology and usage of such technologies has provided epitopes for creating chimeric, multi-epitope vaccine for several organisms like Hepatitis B Virus or Escherichia coli [23]. Proof of identity of B-cell epitopes acting an important role in development of epitopebased vaccines, therapeutic antibodies, and diagnostic tools and T-cell epitopes are shares of intracellular dealing out antigens that are presented to T lymphocytes in association with molecules of the key histocompatibility complex [23,24].
Syphilis vaccination studies are still in the elementary/preclinical stage. The aim of this study is to apply immunoinformatics on β-barrel outer membrane protein to design a safe and effective vaccine. No previous reports were found so this may be considered the first study to use in silico approach to design an epitope-based vaccine [25].
Protein sequence retrieval
A total of 66 Treponema pallidum outer membrane beta-barrel protein strains were retrieved from National Center for Biotechnology Information (NCBI) database on July 2019 in FASTA format. These strains were obtained from different parts of the world for immunoinformatics analysis. The retrieved protein strains had length of 219 amino acid.
Determination of conserved regions
The retrieved sequences of Treponema pallidum outer membrane beta-barrel protein were subjected to multiple sequence alignment (MSA) by blasting them against reference sequence (WP_010882178.1) using ClustalW tool of BioEdit Sequence Alignment Editor Software version 7.2-0.5 to determine the conserved regions. Peptides allocated at highly conserved regions will most likely develop in to stronger vaccine that covers more populations. The molecular weight and amino acid composition of the protein were also retrieved [26].
Sequenced-based method
The reference sequence of Treponema pallidum outer membrane beta-barrel protein was submitted to different prediction tools at the Immune Epitope Database (IEDB) analysis resource (http://www.iedb.org/) to predict various B and T cell epitopes. Conserved epitopes would be considered as candidate epitopes for B and T cells [27].
B cell epitope prediction
B cell epitopes is the part of the vaccine that interacts with B-lymphocytes. Candidate epitopes were analysed using several B cell prediction methods from IEDB (http://tools.iedb.org/bcell/), to identify the surface accessibility, antigenicity and hydrophilicity with the aid of computerized algorithm. Peptides with length larger than eight peptides were spliced to increase the possibility of obtaining peptides with higher scores in the tests. The Bepipred Linear Epitope Prediction 2 was used to predict linear B cell epitope with default threshold value 0.500 (http://tools.iedb.org/bcell/ result/). The Emini Surface Accessibility Prediction tool was used to detect the surface accessibility with default threshold value 1.000 (http://tools.iedb.org/bcell/result/). The kolaskar and tongaonkar antigenicity method was used to identify the antigenicity sites of candidate epitope with default threshold value 1.018 (http://tools. iedb.org/bcell/result/). Parker Hydrophilicity Prediction tool was used to identify the hydrophilic, accessible, or mobile regions with default threshold value 1.447 [28-31].
T cell epitope prediction MHC Class I binding
T cell epitopes is the part of the vaccine that interacts with T lymphocytes. Analysis of peptide binding to the MHC (Major Histocompatibility complex) class I molecule was assessed by the IEDB MHC I prediction tool (http://tools.iedb.org/mhci/). Artificial Neural Network (ANN) 4.0 prediction method was used to predict the binding affinity. Before the prediction, all human allele lengths were selected and set to nine aminoacids. The halfmaximal inhibitory concentration (IC50) value required for all conserved epitopes to bind at score less than or equal to 100 were selected [32-34].
T cell epitope prediction MHC Class II binding
Prediction of T cell epitopes interacting with MHC Class II was assessed by the IEDB MHC II prediction tool (http://tools.iedb. org/mhcii/). There are six available tools for prediction: SMM_ align, NN-align, Combinatorial Libraries, Sturniolo's method, Net MHCII pan and consensus method. Human allele references set were used to determine the interaction potentials of T cell epitopes and MHC Class II allele (HLA DR, DP and DQ). NN-align method was used to predict the binding affinity. All human allele lengths were set to the standard length. IC50 values at score less than or equal to 500 were selected [35,36].
Population coverage
In IEDB, the population coverage link was selected to analyse the epitopes. This tool calculates the fraction of individuals predicted to respond to a given set of epitopes with known MHC restrictions (http://tools.iedb.org/population/iedbinput). The appropriate checkbox for calculation was checked based on MHC I, MHC II separately and combination of both which is set against the whole world population [37].
AllerTOP v. 2.0
This method is used for allergenicity predictions; it is based on auto cross covariance (ACC) transformation of protein sequences into uniform equal-length vectors. The reference protein sequence (WP_010882178.1) was used. The principal Characteristics of the amino acids were represented by five E descriptors, which indicate amino acid hydrophobicity, molecular size, helix-forming propensity, relative abundance of amino acids, and β-strand forming propensity.
Homology modeling
The 3D structure was obtained using raptorX which is a protein structure prediction server developed by Xu group, excelling at predicting secondary and tertiary structure for protein sequences without close homologs in the Protein Data Bank (PDB). Obtained 3D protein structure was visualized by USCF chimera (version 1.8) which was also used for visualization and analysis of molecular structure of the promising epitopes and it’s binding to MHC class I, MHC class II [38,39].
Multiple sequence alignment
The conserved regions and amino acid composition for the reference sequence of Treponeman Pallidum Outer Membrane Beta Barrel Protein are illustrated in Figures 1 and 2 respectively. Glycine, serine and leucine were the most abundant amino acids (Table 1).
| Amino Acid | Number | Mol% |
|---|---|---|
| Ala | 18 | 8.22 |
| Cys | 4 | 1.83 |
| Asp | 5 | 2.28 |
| Glu | 6 | 2.74 |
| Phe | 9 | 4.11 |
| Gly | 29 | 13.24 |
| His | 2 | 0.91 |
| Ile | 12 | 5.48 |
| Lys | 6 | 2.74 |
| Leu | 19 | 8.68 |
| Met | 1 | 0.46 |
| Asn | 9 | 4.11 |
| Pro | 13 | 5.94 |
| Gln | 9 | 4.11 |
| Arg | 9 | 4.11 |
| Ser | 21 | 9.59 |
| Thr | 13 | 5.94 |
| Val | 16 | 7.31 |
| Trp | 3 | 1.37 |
| Tyr | 15 | 6.85 |
Table 1: Molecular weight and amino acid frequency distribution of the protein.

Figure 1: Multiple Sequence Alignment using BioEdit software.
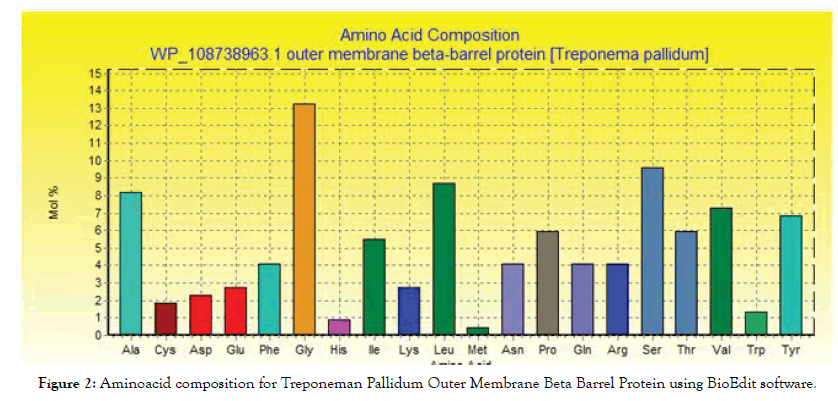
Figure 2: Aminoacid composition for Treponeman Pallidum Outer Membrane Beta Barrel Protein using BioEdit software.
B-cell epitope prediction
The reference sequence of Treponeman Pallidum Outer Membrane Beta Barrel Protein was subjected to Bepipred linear epitope 2, Emini surface accessibility, Kolaskar and Tongaonkar antigenicity and Parker hydrophilicity prediction methods to test for various immunogenicity parameters (Table 2 and Figures 3-8). Two epitopes have successfully passed the three tests. Three dimensional structure of the proposed B cell epitopes is shown in Figures 7 and 8.
| Peptide | Start | End | Length | Emini surface accessibility score (TH: 1) | Kolaskar and Tongaonkar antigenicity score (TH: 1.044) | Parker Hydrophilicity prediction score (TH: 1.196) | ALLERTOP |
|---|---|---|---|---|---|---|---|
| LPAYSSEGVREVPPSQ | 21 | 36 | 16 | 3.52 | 1.058 | 2.556 | Non Allergen |
| SSEGVREVPPSQSPQV | 25 | 40 | 16 | 4.645 | 1.054 | 3.669 | Non Allergen |
| EGVREVPPSQSPQVVV | 27 | 42 | 16 | 1.425 | 1.101 | 2.394 | Non Allergen |
| GQVSFECYRTT | 110 | 120 | 11 | 1.091 | 1.045 | 2.473 | Non Allergen |
| QVSFECYRTTG | 111 | 121 | 11 | 1.091 | 1.045 | 2.473 | Non Allergen |
| VSFECYRTTGSNYYF | 112 | 126 | 15 | 1.503 | 1.045 | 1.447 | Non Allergen |
| FECYRTTGSNYYFSV | 114 | 128 | 15 | 1.503 | 1.045 | 1.447 | Non Allergen |
| GVGLNIQSYLSKKAPP* | 150 | 165 | 16 | 1.101 | 1.05 | 1.4 | Non Allergen |
| AEASAGLYYQYTP | 168 | 180 | 13 | 2.611 | 1.05 | 1.9 | Non Allergen |
| SAGLYYQYTPD* | 171 | 181 | 11 | 3.418 | 1.049 | 2.064 | Non Allergen |
Table 2: List of conserved epitopes that has successfully passed the four tests along with their relative scores. *Proposed epitopes.
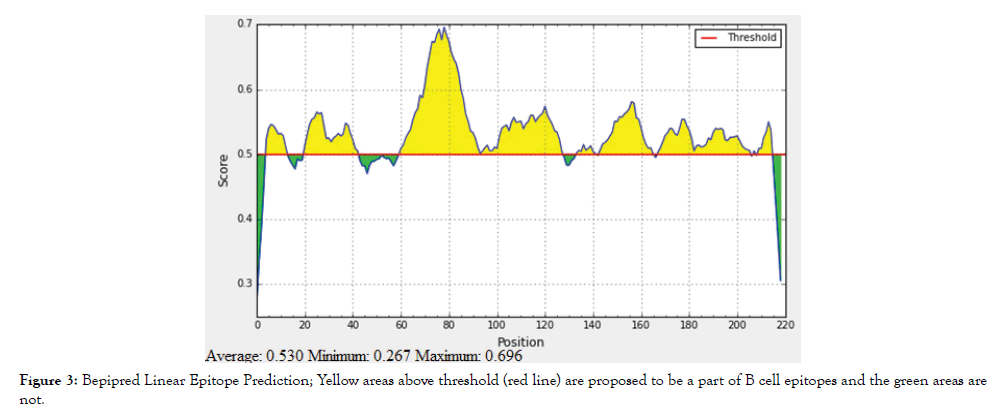
Figure 3: Bepipred Linear Epitope Prediction; Yellow areas above threshold (red line) are proposed to be a part of B cell epitopes and the green areas are not.
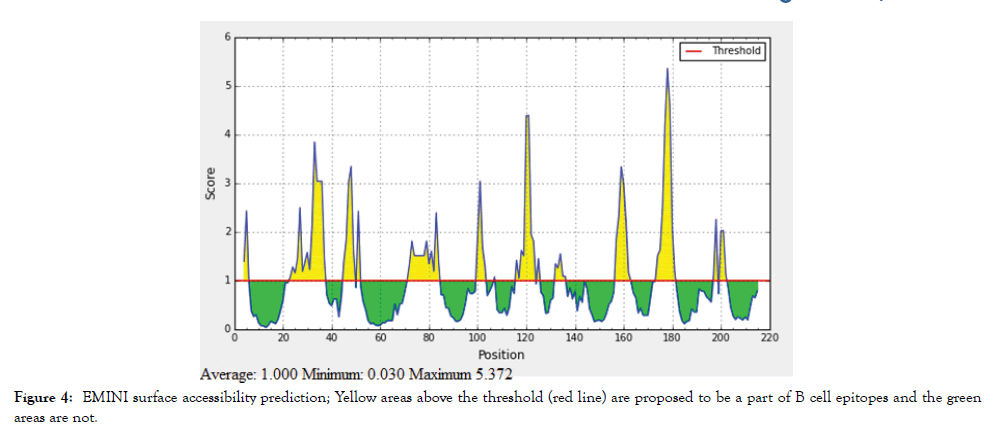
Figure 4: EMINI surface accessibility prediction; Yellow areas above the threshold (red line) are proposed to be a part of B cell epitopes and the green areas are not.
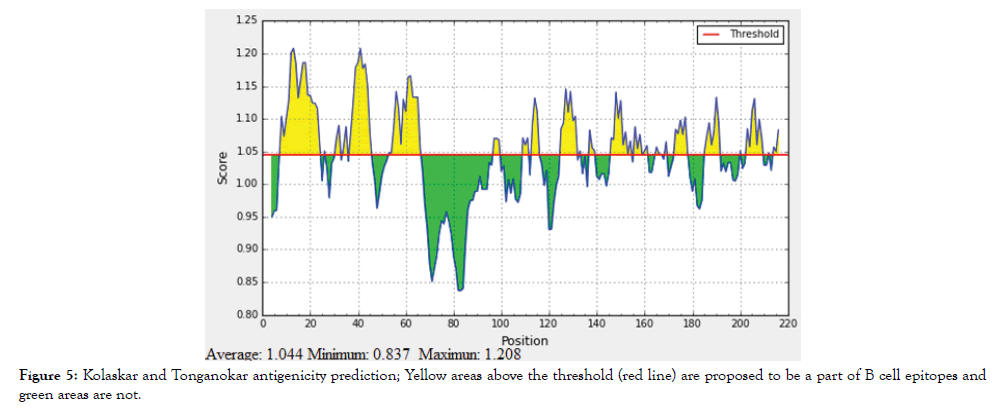
Figure 5: Kolaskar and Tonganokar antigenicity prediction; Yellow areas above the threshold (red line) are proposed to be a part of B cell epitopes and green areas are not.
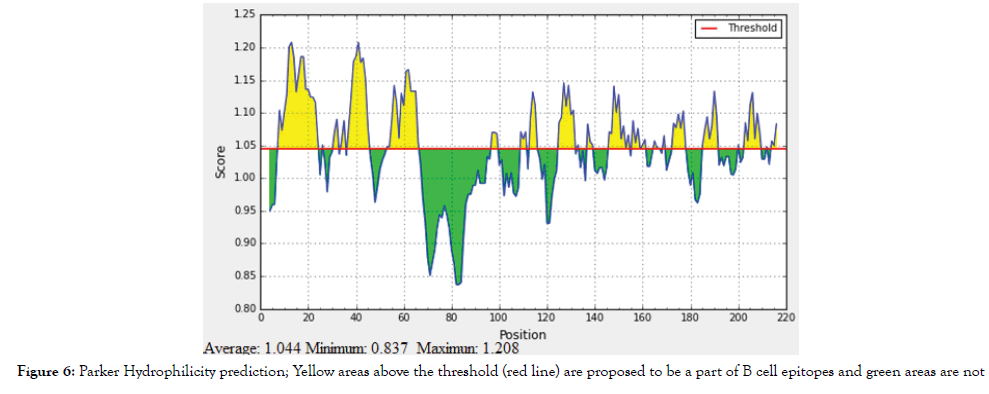
Figure 6: Parker Hydrophilicity prediction; Yellow areas above the threshold (red line) are proposed to be a part of B cell epitopes and green areas are not.
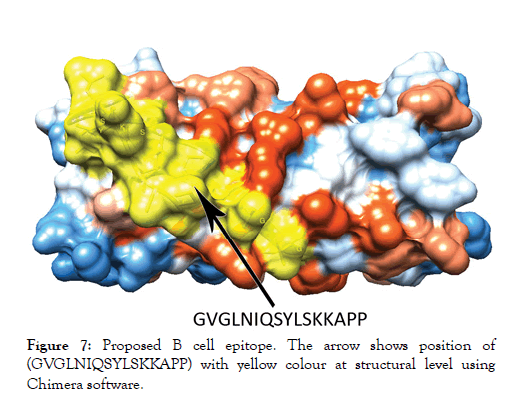
Figure 7: Proposed B cell epitope. The arrow shows position of (GVGLNIQSYLSKKAPP) with yellow colour at structural level using Chimera software.
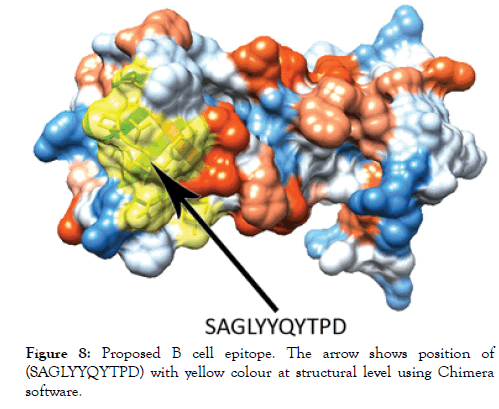
Figure 8: Proposed B cell epitope. The arrow shows position of (SAGLYYQYTPD) with yellow colour at structural level using Chimera software.
Prediction of cytotoxic T-lymphocyte epitopes and modeling
The reference sequence was analyzed using (IEDB) MHC-1 binding prediction tool to predict T cell epitopes interacting with different types of MHC Class I alleles, based on Artificial Neural Network (ANN) with half-maximal inhibitory concentration (IC50)<100 nm. 36 peptides were predicted to interact with different MHC-1alleles. The most promising epitopes and their corresponding MHC-1 alleles are shown in Table 3. The tertiary structure of the promising epitopes was missing in the homology model.
| Peptide | MHC 1 alleles |
|---|---|
| AEASAGLYY | HLA-B*44:03, HLA-B*44:02, HLA-B*15:01, HLA-A*29:02 |
| ALCALSPLL | HLA-A*02:01 |
| ALSPLLPAY | HLA-B*15:01, HLA-A*30:02, HLA-A*29:02 |
| ASAGLYYQY | HLA-A*29:02, HLA-A*30:02, HLA-B*58:01, HLA-A*11:01 |
| CALSPLLPA | HLA-A*02:06 |
| CYRTTGSNY | HLA-C*14:02 |
| FSLGGQVSF | HLA-C*03:03, HLA-B*35:01 |
| GALCALSPL | HLA-A*02:06 |
| GQVSFECYR | HLA-A*31:01 |
| GRWRIPLSL | HLA-B*27:05 |
| IAEASAGLY | HLA-A*30:02 |
| IPLSLGVGL | HLA-B*07:02 |
| ITVNPTYTF | HLA-A*32:01, HLA-B*58:01, HLA-B*57:01, HLA-A*23:01 |
| LGYEYFFTK | HLA-A*11:01 |
| LIAEASAGL | HLA-A*02:06, HLA-A*68:02 |
| LYYQYTPDW | HLA-C*14:02, HLA-A*23:01, HLA-A*24:02 |
| MSRTFRAWQ | HLA-A*30:01 |
| NIQSYLSKK | HLA-A*11:01, HLA-A*68:01 |
| QCVGALCAL | HLA-C*03:03 |
| QVVVAYEPI | HLA-A*68:02, HLA-A*02:06 |
| RAWQCVGAL | HLA-B*07:02, HLA-C*03:03, HLA-C*12:03 |
| RTFRAWQCV | HLA-A*30:01, HLA-A*02:06, HLA-A*32:01, HLA-C*15:02, HLA-C*12:03 |
| RTTGSNYYF | HLA-B*58:01, HLA-B*57:01, HLA-A*32:01 |
| SNYYFSVPI | HLA-A*32:01, HLA-A*68:02 |
| TVNPTYTFA | HLA-A*68:02 |
| VAYEPIRPG | HLA-C*12:03 |
| VPITVNPTY | HLA-B*35:01, HLA-B*53:01 |
| WRIPLSLGV | HLA-B*27:05 |
| WSIGGIVAY | HLA-B*35:01, HLA-A*29:02, HLA-C*12:03, HLA-B*15:01 |
| YEYFFTKNF | HLA-B*18:01, HLA-C*12:03, HLA-B*44:03, HLA-B*40:02, HLA-C*14:02, HLA-B*44:02 |
| YFFTKNFSL | HLA-C*14:02, HLA-C*03:03, HLA-B*08:01 |
| YLSKKAPGL | HLA-A*02:01 |
| YQYTPDWSI | HLA-A*02:06, HLA-A*02:01, HLA-C*03:03, HLA-B*39:01, HLA-C*12:03 |
| YRTTGSNYY | HLA-C*06:02, HLA-C*07:01 |
| YSSEGVREV | HLA-C*12:03 |
| YYFSVPITV | HLA-C*12:03, HLA-C*14:02, HLA-A*23:01, HLA-A*24:02, HLA-C*07:02 |
Table 3: The most promising T cell epitopes and their corresponding MHC-1 alleles.
Prediction of the helper T-lymphocyte epitopes and modeling
Reference sequence was analyzed using (IEDB) MHC-II binding prediction tool based on NN-align with half-maximal inhibitory concentration (IC50)<500 nm; there were 135 predicted epitopes found to interact with MHC-II alleles. The most promising epitopes and their corresponding alleles are shown in Table 4 along with the 3D structure of the proposed epitope (Figure 9).
| Peptide | MHC 2 alleles |
|---|---|
| SNYYFSVPITVNPTY | HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*04:01, HLA- DPA1*03:01/DPB1*04:02, HLA-DRB1*15:01, HLA-DPA1*02:01/DPB1*01:01, HLA- DRB1*04:05, HLA-DRB1*04:04, HLA-DRB1*13:02, HLA-DRB1*01:01, HLA- DQA1*05:01/DQB1*03:01, HLA-DRB1*07:01, HLA-DQA1*01:01/DQB1*05:01, HLA- DQA1*01:02/DQB1*06:02, HLA-DRB1*08:02, HLA-DPA1*01/DPB1*04:01 QVVVAYEPIRPGDQL HLA-DRB1*15:01, HLA-DRB4*01:01, HLA-DRB1*01:01, HLA-DRB1*08:02, HLA-DRB1*04:04, HLA-DPA1*03:01/DPB1*04:02, HLA- DQA1*05:01/DQB1*02:01 |
| GSNYYFSVPITVNPT | HLA-DRB1*09:01, HLA-DRB1*11:01, HLA-DRB1*15:01, HLA-DRB1*04:01, HLA- DPA1*03:01/DPB1*04:02, HLA-DRB1*04:05, HLA-DPA1*02:01/DPB1*01:01, HLA- DRB1*13:02, HLA-DRB1*04:04, HLA-DRB1*01:01, HLA-DQA1*05:01/DQB1*03:01, HLA-DRB1*07:01, HLA-DQA1*01:01/DQB1*05:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DRB5*01:01, HLA-DPA1*01/DPB1*04:01 SIGGIVAYTQLGDIA HLA-DPA1*02:01/DPB1*01:01, HLA- DPA1*01:03/DPB1*02:01, HLA-DRB1*15:01, HLA-DRB1*07:01, HLA-DRB1*04:04, HLA-DQA1*05:01/DQB1*03:01, HLA-DRB1*01:01, HLA-DRB4*01:01, HLA- DRB1*04:05, HLA-DPA1*03:01/DPB1*04:02, HLA-DRB1*08:02 |
| EYFFTKNFSLGGQVS | HLA-DRB5*01:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*09:01, HLA- DPA1*02:01/DPB1*01:01, HLA-DRB1*04:05, HLA-DRB1*15:01, HLA- DPA1*03:01/DPB1*04:02, HLA-DRB1*01:01, HLA-DRB1*11:01, HLA-DRB1*07:01, HLA-DRB1*04:01, HLA-DPA1*01/DPB1*04:01, HLA-DPA1*02:01/DPB1*05:01, HLA- DQA1*05:01/DQB1*03:01, HLA-DRB1*13:02, HLA-DRB1*04:04 |
| YFFTKNFSLGGQVSF | HLA-DRB1*09:01, HLA-DRB1*04:01, HLA-DPA1*01/DPB1*04:01, HLA-DRB5*01:01, HLA-DQA1*05:01/DQB1*03:01, HLA-DPA1*01:03/DPB1*02:01, HLA- DPA1*02:01/DPB1*01:01, HLA-DRB1*04:05, HLA-DRB1*01:01, HLA-DRB1*11:01, HLA-DRB1*07:01, HLA-DRB1*15:01, HLA-DPA1*03:01/DPB1*04:02, HLA- DRB1*04:04, HLA-DRB1*13:02 |
| LGYEYFFTKNFSLGG | HLA-DRB1*04:05, HLA-DPA1*02:01/DPB1*05:01, HLA-DRB1*15:01, HLA- DRB1*04:04, HLA-DRB1*09:01, HLA-DPA1*03:01/DPB1*04:02, HLA-DRB1*01:01, HLA-DRB1*07:01, HLA-DPA1*01/DPB1*04:01, HLA-DRB5*01:01, HLA- DPA1*02:01/DPB1*01:01, HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*04:01, HLA- DRB1*11:01, HLA-DQA1*01:01/DQB1*05:01 |
Table 4: The most promising T cell epitopes and their corresponding MHC-2 alleles.
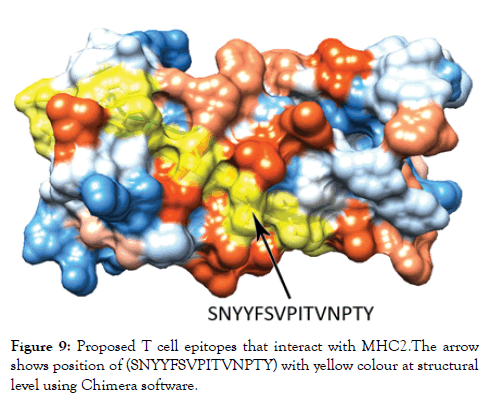
Figure 9: Proposed T cell epitopes that interact with MHC2. The arrow shows position of (SNYYFSVPITVNPTY) with yellow colour at structural level using Chimera software.
Population coverage analysis
All MHC I and MHC II epitopes were assessed for population coverage against the whole world using IEDB population coverage tool. For MHC 1, epitopes with highest population coverage were YQYTPDWSI (52.63) and YYFSVPITV (50.62) (Figure 10 and Table 5). For MHC class II, the epitopes that showed highest population coverage were SNYYFSVPITVNPTY (73.11), GSNYYFSVPIT VNPT and EYFFTKNFSLGGQVS (71.88%) (Tables 6-9). When combined together, the epitopes that showed highest population coverage were SNYYFSVPITVNPTY (73.11), GSNYYFSVPITVNPT and EYFFTKNFSLGGQVS (71.88%) (Figure 11, Tables 7 and 8).
| Epitope | Coverage Class 1 (%) | Total HLA hits |
|---|---|---|
| YQYTPDWSI | 0.5263 | 5 |
| YYFSVPITV | 0.5062 | 5 |
| ALCALSPLL | 0.3908 | 1 |
| YLSKKAPGL | 0.3908 | 1 |
| YEYFFTKNF | 0.3359 | 6 |
| YRTTGSNYY | 0.3331 | 2 |
| RAWQCVGAL | 0.2847 | 3 |
| LYYQYTPDW | 0.2843 | 3 |
| WSIGGIVAY | 0.2801 | 4 |
| AEASAGLYY | 0.2452 | 4 |
| ASAGLYYQY | 0.2397 | 4 |
| RTFRAWQCV | 0.2327 | 5 |
| NIQSYLSKK | 0.2088 | 2 |
| YFFTKNFSL | 0.2042 | 3 |
| ITVNPTYTF | 0.1645 | 4 |
| FSLGGQVSF | 0.1585 | 2 |
| LGYEYFFTK | 0.1553 | 1 |
| ALSPLLPAY | 0.1418 | 3 |
| IPLSLGVGL | 0.1278 | 1 |
| RTTGSNYYF | 0.1153 | 3 |
| VPITVNPTY | 0.1084 | 2 |
| VAYEPIRPG | 0.1031 | 1 |
| YSSEGVREV | 0.1031 | 1 |
| QCVGALCAL | 0.0812 | 1 |
| SNYYFSVPI | 0.0705 | 2 |
| GQVSFECYR | 0.0536 | 1 |
| GRWRIPLSL | 0.0478 | 1 |
| WRIPLSLGV | 0.0478 | 1 |
| LIAEASAGL | 0.0443 | 2 |
| QVVVAYEPI | 0.0443 | 2 |
Table 5: Population coverage of proposed peptides interaction with MHC class I.
| Epitope | Coverage Class 2 (%) | Total HLA hits |
|---|---|---|
| SNYYFSVPITVNPTY | 0.7311 | 16 |
| GSNYYFSVPITVNPT | 0.7188 | 16 |
| EYFFTKNFSLGGQVS | 0.7188 | 16 |
| YFFTKNFSLGGQVSF | 0.7188 | 15 |
| YEYFFTKNFSLGGQV | 0.7188 | 15 |
| TGSNYYFSVPITVNP | 0.7188 | 14 |
| IPLSLGVGLNIQSYL | 0.7024 | 11 |
| TTGSNYYFSVPITVN | 0.6926 | 13 |
| LNIQSYLSKKAPGLI | 0.6887 | 10 |
| LGYEYFFTKNFSLGG | 0.6815 | 15 |
| GYEYFFTKNFSLGGQ | 0.6815 | 14 |
| PLSLGVGLNIQSYLS | 0.6776 | 11 |
| LSLGVGLNIQSYLSK | 0.6776 | 11 |
| ITVNPTYTFAVGRWR | 0.6755 | 10 |
| NIQSYLSKKAPGLIA | 0.6755 | 9 |
| IQSYLSKKAPGLIAE | 0.6755 | 9 |
| GLNIQSYLSKKAPGL | 0.6496 | 9 |
| HLGYEYFFTKNFSLG | 0.6437 | 14 |
| RIPLSLGVGLNIQSY | 0.6404 | 10 |
| FFTKNFSLGGQVSFE | 0.6356 | 8 |
| FTKNFSLGGQVSFEC | 0.6356 | 8 |
| YFSVPITVNPTYTFA | 0.6298 | 10 |
| RTTGSNYYFSVPITV | 0.6296 | 9 |
| NYYFSVPITVNPTYT | 0.6214 | 13 |
| YYFSVPITVNPTYTF | 0.6214 | 13 |
| SAGLYYQYTPDWSIG | 0.6174 | 15 |
| AGLYYQYTPDWSIGG | 0.6174 | 14 |
| LYYQYTPDWSIGGIV | 0.6174 | 14 |
| ASAGLYYQYTPDWSI | 0.6174 | 14 |
| GLYYQYTPDWSIGGI | 0.6174 | 13 |
| QVSFECYRTTGSNYY | 0.6174 | 10 |
| VSFECYRTTGSNYYF | 0.6174 | 9 |
| SFECYRTTGSNYYFS | 0.6174 | 9 |
| FECYRTTGSNYYFSV | 0.6174 | 9 |
| FSVPITVNPTYTFAV | 0.6111 | 9 |
| SVPITVNPTYTFAVG | 0.6111 | 8 |
| PGLIAEASAGLYYQY | 0.6109 | 10 |
| APGLIAEASAGLYYQ | 0.6109 | 10 |
| YTFAVGRWRIPLSLG | 0.6086 | 13 |
| TFAVGRWRIPLSLGV | 0.6086 | 12 |
| VGRWRIPLSLGVGLN | 0.6086 | 9 |
| FAVGRWRIPLSLGVG | 0.6086 | 8 |
| AVGRWRIPLSLGVGL | 0.6086 | 8 |
| QSYLSKKAPGLIAEA | 0.6064 | 9 |
| WRIPLSLGVGLNIQS | 0.5984 | 7 |
| TYTFAVGRWRIPLSL | 0.5939 | 12 |
| GLIAEASAGLYYQYT | 0.5672 | 10 |
| KAPGLIAEASAGLYY | 0.5672 | 8 |
| KKAPGLIAEASAGLY | 0.5672 | 8 |
| SKKAPGLIAEASAGL | 0.5672 | 7 |
| NPTYTFAVGRWRIPL | 0.5625 | 9 |
| VNPTYTFAVGRWRIP | 0.5625 | 9 |
| TVNPTYTFAVGRWRI | 0.5625 | 8 |
| GRWRIPLSLGVGLNI | 0.5625 | 7 |
| LGVGLNIQSYLSKKA | 0.5597 | 11 |
| PTYTFAVGRWRIPLS | 0.5513 | 10 |
| VGLNIQSYLSKKAPG | 0.5271 | 8 |
| GVGLNIQSYLSKKAP | 0.5271 | 8 |
| SLGVGLNIQSYLSKK | 0.5154 | 10 |
| SIGGIVAYTQLGDIA | 0.5141 | 11 |
| WSIGGIVAYTQLGDI | 0.5141 | 11 |
| GALCALSPLLPAYSS | 0.5064 | 13 |
| VGALCALSPLLPAYS | 0.5064 | 13 |
| ALCALSPLLPAYSSE | 0.5064 | 12 |
| LCALSPLLPAYSSEG | 0.5014 | 10 |
| QCVGALCALSPLLPA | 0.4899 | 12 |
| WQCVGALCALSPLLP | 0.4884 | 11 |
| YYQYTPDWSIGGIVA | 0.4884 | 9 |
| LIAEASAGLYYQYTP | 0.4879 | 9 |
| RWRIPLSLGVGLNIQ | 0.4879 | 6 |
| SYLSKKAPGLIAEAS | 0.4757 | 8 |
| CVGALCALSPLLPAY | 0.4679 | 11 |
| CYRTTGSNYYFSVPI | 0.4628 | 7 |
| YRTTGSNYYFSVPIT | 0.4628 | 7 |
| MSRTFRAWQCVGALC | 0.4627 | 6 |
| FRAWQCVGALCALSP | 0.4612 | 8 |
| RTFRAWQCVGALCAL | 0.4612 | 7 |
| TFRAWQCVGALCALS | 0.4612 | 7 |
| SRTFRAWQCVGALCA | 0.4612 | 7 |
| SPQVVVAYEPIRPGD | 0.4527 | 10 |
| VPITVNPTYTFAVGR | 0.4504 | 6 |
| PQVVVAYEPIRPGDQ | 0.4478 | 10 |
| ECYRTTGSNYYFSVP | 0.4403 | 5 |
| YQYTPDWSIGGIVAY | 0.4306 | 7 |
| CALSPLLPAYSSEGV | 0.422 | 8 |
| RAWQCVGALCALSPL | 0.4124 | 7 |
| AWQCVGALCALSPLL | 0.4124 | 7 |
| ALSPLLPAYSSEGVR | 0.404 | 7 |
| YLSKKAPGLIAEASA | 0.3943 | 7 |
| LSPLLPAYSSEGVRE | 0.3803 | 5 |
| PITVNPTYTFAVGRW | 0.3615 | 5 |
| DWSIGGIVAYTQLGD | 0.3527 | 7 |
| EASAGLYYQYTPDWS | 0.35 | 7 |
| GQVSFECYRTTGSNY | 0.3484 | 6 |
| QVVVAYEPIRPGDQL | 0.3471 | 7 |
| VVVAYEPIRPGDQLL | 0.3471 | 4 |
| AVGLATIDFGVRYHF | 0.3444 | 3 |
| TKNFSLGGQVSFECY | 0.3402 | 5 |
| KNFSLGGQVSFECYR | 0.3402 | 5 |
| QSPQVVVAYEPIRPG | 0.3055 | 8 |
| SQSPQVVVAYEPIRP | 0.2858 | 5 |
| SPLLPAYSSEGVREV | 0.2187 | 5 |
| PIRPGDQLLKIGIVA | 0.2143 | 3 |
| IRPGDQLLKIGIVAG | 0.2143 | 2 |
| YEPIRPGDQLLKIGI | 0.1784 | 1 |
| LSKKAPGLIAEASAG | 0.1782 | 4 |
| TPDWSIGGIVAYTQL | 0.1755 | 5 |
| NFSLGGQVSFECYRT | 0.1755 | 5 |
| YTPDWSIGGIVAYTQ | 0.1755 | 4 |
| QYTPDWSIGGIVAYT | 0.1755 | 4 |
| LGGQVSFECYRTTGS | 0.157 | 4 |
| GGQVSFECYRTTGSN | 0.157 | 4 |
| PDWSIGGIVAYTQLG | 0.1153 | 3 |
| AYEPIRPGDQLLKIG | 0.1153 | 1 |
| VAYEPIRPGDQLLKI | 0.1153 | 1 |
| VVAYEPIRPGDQLLK | 0.1153 | 1 |
| AEASAGLYYQYTPDW | 0.0771 | 4 |
| FSLGGQVSFECYRTT | 0.064 | 5 |
| IAEASAGLYYQYTPD | 0 | 4 |
| SLGGQVSFECYRTTG | 0 | 2 |
| PLLPAYSSEGVREVP | 0 | 2 |
| LPAYSSEGVREVPPS | 0 | 2 |
| VPPSQSPQVVVAYEP | 0 | 2 |
| PPSQSPQVVVAYEPI | 0 | 2 |
| LLPAYSSEGVREVPP | 0 | 2 |
| SEGVREVPPSQSPQV | 0 | 1 |
| PAYSSEGVREVPPSQ | 0 | 1 |
| PSQSPQVVVAYEPIR | 0 | 1 |
| GVREVPPSQSPQVVV | 0 | 1 |
| EPIRPGDQLLKIGIV | 0 | 1 |
| SSEGVREVPPSQSPQ | 0 | 1 |
| EGVREVPPSQSPQVV | 0 | 1 |
| YSSEGVREVPPSQSP | 0 | 1 |
| AYSSEGVREVPPSQS | 0 | 1 |
| VREVPPSQSPQVVVA | 0 | 1 |
Table 6: Population coverage of proposed peptides interaction with MHC class 2.
| Epitope | Coverage Class 1and2 combined (%) | Total HLA hits |
|---|---|---|
| SNYYFSVPITVNPTY | 0.7311 | 16 |
| GSNYYFSVPITVNPT | 0.7188 | 16 |
| EYFFTKNFSLGGQVS | 0.7188 | 16 |
| YFFTKNFSLGGQVSF | 0.7188 | 15 |
| YEYFFTKNFSLGGQV | 0.7188 | 15 |
| TGSNYYFSVPITVNP | 0.7188 | 14 |
| IPLSLGVGLNIQSYL | 0.7024 | 11 |
| TTGSNYYFSVPITVN | 0.6926 | 13 |
| LNIQSYLSKKAPGLI | 0.6887 | 10 |
| LGYEYFFTKNFSLGG | 0.6815 | 15 |
| GYEYFFTKNFSLGGQ | 0.6815 | 14 |
| PLSLGVGLNIQSYLS | 0.6776 | 11 |
| LSLGVGLNIQSYLSK | 0.6776 | 11 |
| ITVNPTYTFAVGRWR | 0.6755 | 10 |
| NIQSYLSKKAPGLIA | 0.6755 | 9 |
| IQSYLSKKAPGLIAE | 0.6755 | 9 |
| GLNIQSYLSKKAPGL | 0.6496 | 9 |
| HLGYEYFFTKNFSLG | 0.6437 | 14 |
| RIPLSLGVGLNIQSY | 0.6404 | 10 |
| FFTKNFSLGGQVSFE | 0.6356 | 8 |
| FTKNFSLGGQVSFEC | 0.6356 | 8 |
| YFSVPITVNPTYTFA | 0.6298 | 10 |
| RTTGSNYYFSVPITV | 0.6296 | 9 |
| NYYFSVPITVNPTYT | 0.6214 | 13 |
| YYFSVPITVNPTYTF | 0.6214 | 13 |
| SAGLYYQYTPDWSIG | 0.6174 | 15 |
| AGLYYQYTPDWSIGG | 0.6174 | 14 |
| LYYQYTPDWSIGGIV | 0.6174 | 14 |
| ASAGLYYQYTPDWSI | 0.6174 | 14 |
| GLYYQYTPDWSIGGI | 0.6174 | 13 |
| QVSFECYRTTGSNYY | 0.6174 | 10 |
| VSFECYRTTGSNYYF | 0.6174 | 9 |
| SFECYRTTGSNYYFS | 0.6174 | 9 |
| FECYRTTGSNYYFSV | 0.6174 | 9 |
| FSVPITVNPTYTFAV | 0.6111 | 9 |
| SVPITVNPTYTFAVG | 0.6111 | 8 |
| PGLIAEASAGLYYQY | 0.6109 | 10 |
| APGLIAEASAGLYYQ | 0.6109 | 10 |
| YTFAVGRWRIPLSLG | 0.6086 | 13 |
| TFAVGRWRIPLSLGV | 0.6086 | 12 |
| VGRWRIPLSLGVGLN | 0.6086 | 9 |
| FAVGRWRIPLSLGVG | 0.6086 | 8 |
| AVGRWRIPLSLGVGL | 0.6086 | 8 |
| QSYLSKKAPGLIAEA | 0.6064 | 9 |
| WRIPLSLGVGLNIQS | 0.5984 | 7 |
| TYTFAVGRWRIPLSL | 0.5939 | 12 |
| GLIAEASAGLYYQYT | 0.5672 | 10 |
Table 7: Population coverage of proposed peptide interaction with MHC class 1 and 2 combined.
| Country | MHC I | MHC II | MHC IandII combined |
|---|---|---|---|
| World | 98.2% | 68.45 % * | 99.64 % * |
Table 8: The population coverage of whole world for the epitope set for MHC I, MHC II and MHC I and II combined.
| Population/ Area | Class II | ||
|---|---|---|---|
| World | Coveragea | Average_hitb | pc90c |
| Average | 68.45% | 53.07 | -1.16 |
| Standard Deviation | 0.0 | 0.0 | 0.0 |
Table 9: Population coverage for MHC class II epitopes.
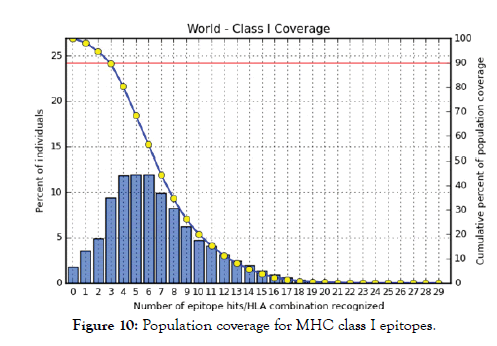
Figure 10: Population coverage for MHC class I epitopes.
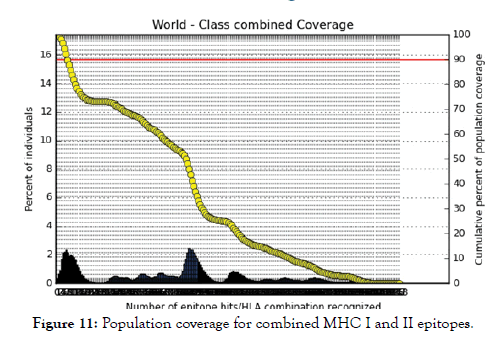
Figure 11: Population coverage for combined MHC I and II epitopes.
In population coverage analysis of MHC II; 14 alleles were not included in the calculation, therefore the above (*) percentages are for epitope sets excluding: HLA-DQA1*05:01/DQB1*03:01, HLA-DQA1*01:02/DQB1*06:02, HLA-DPA1*01:03/DPB1*02:01, HLA-DPA1*01/DPB1*04:01, HLA-DPA1*02:01 /DPB1*05:01, HLA-DQA1*01:01/DQB1*05:01, HLA-DQA1*03:01/DQB1*03:02, HLA-DRB3*01:01, HLA-DRB4*01:01, HLA-DRB5*01:01, HLADQA1* 04:01/DQB1*04:02, HLA-DPA1*03:01/DPB1*04:02, HLADQA1* 05:01/DQB1*02:01, HLA-DPA1*02:01/DPB1*01:01.
In this study, we successfully designed a peptide vaccine for Treponema pallidum against its immunogenic outer membrane beta-barrel protein by immunoinformatics tools. These peptides can be recognized by B cell and T cell to produce antibodies. Predicted peptide vaccine characterized by easy production, stimulating effective immune response and no potential infection possibilities. Peptide vaccines overcome the side effects of conventional vaccines [40].
The reference sequence of Treponema pallidum outer membrane beta-barrel protein was subjected to Bepipred linear epitope prediction 2 test, Emini surface accessibility test, Kolaskar and Tongaonkar antigenicity test and Parker hydrophilicity test in IEDB, to determine the binding to B cell, surface accessibility, antigenicity, the hydrophilicity of the B cell epitope respectively. Out of the ten predicted epitopes using Bepipred 2 test, only two epitopes passed the other three tests (GVGLNIQSYLSKKAPP, SAGLYYQYTPD). The reference sequence was analysed using MHC Iand MHC II binding prediction tools in IEDB to predict T cell epitopes. 36 epitopes were predicted to interact with MHC I alleles with IC50<100. Four of them were most promising and had the affinity to bind the highest number of MHC1 alleles (YEYFFTKNF, YQYTPDWSI, YYFSVPITV, RTFRAWQCV). 135 epitopes predicted to interact with MHC II alleles with IC50<500. Five of them were most promising and had the affinity to bind to the highest number of MHC II alleles (SNYYFSVPITVNPTY, GSNYYFSVPITVNPT, EYFFTKNFSLGGQVS, YFFTKNFSLGGQVSF, LGYEYFFTKNFSLGG).
The best epitope with the highest population coverage for MHC I was YQYTPDWSI with 52.63% in five HLA hits, and the coverage of population set for whole MHC I epitopes was 98.2%. Excluding cer-tain alleles for MHC II, the best epitope was SNYYFSVPITVNPTY scoring 73.11% with 16 HLA hits and the coverage of population set was 68.45% for the whole Helper Tlymphocyte epitopes (MHC II). In population coverage analysis of MHC II; 14 alleles were not included in the calculation: HLA-DQA1*05:01/DQB1*03:01, HLA-DQA1*01:02/ DQB1*06:02, HLA-DPA1*01:03/DPB1*02:01, HLA-DPA1*01/ DPB1*04:01,HLA-DPA1*02:01/DPB1*05:01,HLA-DQA1*01:01/ DQB1*05:01, HLA-DQA1*03:01/DQB1*03:02, HLADRB3* 01:01, HLA-DRB4*01:01, HLA-DRB5*01:01, HLADQA1* 04:01/DQB1*04:02,HLA-DPA1*03:01/DPB1*04:02, HLA-DQA1*05:01/DQB1*02:01, HLA-DPA1*02:01/DPB1*01:01. These epitopes could potentially induce a T-cell immune response when interacting strongly with MHC I and MHC II alleles effectively generating a cellular immune response against the invading patho-gen. The peptide SNYYFSVPITVNPTY had the highest popula-tion coverage per cent 73.1% in 16 HLA hits for both MHC I and MHC II. Many studies have used immunoinformatics methods to predict peptide vaccines for various micro-organisms such as HPV, Rubella, Dengue, Zika, Ebola, and mycetoma [41-49]. We hope that the whole world will benefit from this epitope-based vaccine upon its successful development following in vivo and in vitro studies to prove its effectiveness.
The method to protect and minimize the chance of infection is known as vaccination.. Vaccines Design by in silico prediction methods is very appreciated due to the important reduction in cost, effort and time. Peptide vaccines overcome the side effects of conventional vaccines. For the first time we designed different peptides that can create antibodies against β -Barrel outer membrane protein of Treponema pallidum. Two B cell epitopes passed the antigenicity, accessibility and hydrophilicity tests. 36 MHC I epitopes were the most promising ones, while five epitopes for MHC II. Five epitopes were common between MHC I and II. For the population coverage, the epitopes covered 71.88% of the alleles worldwide without certain alleles.
The data which support our findings in this study are available from the corresponding author upon reasonable request.
The authors declare that they have no competing interests.
Many thank goes to National Center for Biotechnology Information (NCBI), BioEdit, Immune Epitope Database Analysis Resource (IEDB), RaptorX server, and UCSF Chimera team. We are grateful to Editor-in-Chief and all the unknown reviewers for their valuable comments on the previous version of this manuscript.
Citation: Elhag M, Saadaldin MM, Abdelmoneim AH, Taha TS, Abdelrahman F, Hassan MA (2020) Immunoinformatics Approach for Designing an Epitope-Based Peptide Vaccine against Treponema pallidum Outer Membrane Beta-Barrel Protein. Immunome Res. 16:175. Doi: 10.35248/1745-7580.19.16.175
Received: 24-Feb-2020 Accepted: 03-Mar-2020 Published: 17-Mar-2020 , DOI: 10.35248/1745-7580.20.16.175
Copyright: © 2020 Elhag M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.