Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2014) Volume 7, Issue 6
Formaldehyde-fixed, paraffin-embedded (FFPE) tissue repositories represent a valuable resource for the retrospective study of disease progression and response to therapy. However, the proteomic analysis of FFPE tissues has been hampered by formaldehyde-induced protein modifications, which reduce protein extraction efficiency and may lead to protein misidentification. Here, we demonstrate the use of heat augmented with high hydrostatic pressure (40,000 psi) as a novel method for the recovery of intact proteins from FFPE tissue. Our laboratory has taken a mechanistic approach to developing improved protein extraction protocols, by first studying the reactions of formaldehyde with proteins and ways to reverse these reactions, then applying this approach to a model system called a “tissue surrogate”, which is a gel formed by treating high concentrations of cytoplasmic proteins with formaldehyde, and finally FFPE mouse liver tissue. Our studies indicate that elevated pressure improves the recovery of proteins from FFPE tissue surrogates by hydrating and promoting solubilization of highly aggregated proteins allowing for the subsequent reversal (by hydrolysis) of formaldehyde-induced protein adducts and cross-links. When FFPE mouse liver was extracted using heat and elevated pressure, there was a 4-fold increase in protein extraction efficiency and up to a 30-fold increase in the number of non-redundant proteins identified by mass spectrometry, compared to matched tissue extracted with heat alone. More importantly, the number of non-redundant proteins identified in the FFPE tissue was nearly identical to that of the corresponding frozen tissue.
Keywords: Antigen Retrieval, FFPE, Formalin-fixed paraffinembedded, High-pressure protein extraction, Mass spectrometry, Proteomics
Proteomic technology has advanced to a state where thousands of proteins can be identified within complex samples [1,2], yet diseasebased studies using fresh or frozen tissues are hampered by the limited availability of specimens for longitudinal clinical investigations. In contrast, tissue archives contain millions of formalin-fixed, paraffinembedded (FFPE) tissues for which the clinical course of disease and response to therapy has been established. Unfortunately, protein modifications by formaldehyde treatment and histological processing [3,4] have frustrated attempts to use FFPE tissues for proteomic analyses due to the difficulty in extracting representative proteins. This limitation has restricted studies of diseases that evolve slowly or for those where the time between treatment and recurrence is long, such as prostate and breast cancer. Coupling the medical history and pathology information from FFPE tissues with proteomic investigations would produce a wealth of practical information on important human diseases.
For liquid chromatography-mass spectrometry (LC/MS)-based proteomics, efficient extraction of high-quality proteins is of key importance. Most current extraction protocols for FFPE proteomics have been adapted from heat-induced antigen retrieval methods, originally developed for immunohistochemistry [1,5,6]. Highly encouraging proteomic studies of FFPE tissues have appeared in the recent literature and there are several comprehensive reviews of this topic [3,7,8]. However these investigations have typically been restricted to minute tissue specimens, such as those obtained by laser capture microdissection. Further, some studies report high rates of false-positive protein identification and are limited to the analysis of tryptic digests of FFPE tissues by LC/MS. To develop improved extraction protocols for FFPE tissue, our laboratory has taken a mechanistic approach, by first studying the reactions of formaldehyde with proteins and ways to reverse these reactions [9], then applying this approach to a model system called a “tissue surrogate”, which is a gel formed by treating high concentrations of cytoplasmic proteins with formaldehyde [10-12], and finally FFPE mouse liver tissue [13].
Understanding the effects of formalin-fixation and histological processing on protein structure
As mentioned previously, the recovery and identification by MS of proteins from FFPE tissue have been hampered by the covalent protein modifications and cross-links that are formed during formaldehyde fixation and subsequent histological processing. Three types of formaldehyde-induced chemical modifications have been identified in proteins and model peptides: (a) methylol (hydroxymethyl) adducts, (b) Schiff’s bases, and (c) stable methylene bridges (cross-links) [14,15]. Methylol adducts and Schiff’s bases are easily reversible; however, when present on primary amines or thiols, principally lysine and cysteine, they can undergo a second reaction with a spatially accessible amino acid to form a cross-link. These partnering amino acids are arginine, asparagine, glutamine, histidine, tryptophan, and tyrosine [15,16]. Protein cross-links have been identified in both model peptides [15] and whole proteins, such as insulin [14]. Additionally, the protein N-terminal amine can be converted to a stable 4-imidazolidione adduct [14] and a Mannich reaction can occur between adducted tyrosine and arginine residues in close spatial proximity [17].
Using model proteins in aqueous solution, our laboratory demonstrated that the majority of protein formaldehyde adducts and cross-links were consistently reversed with mild heating following the removal of excess formaldehyde by dialysis [18]. After fixation in 10% buffered formaldehyde, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) of 1 mg/ml solutions of RNase A showed a mixture of intermolecular cross-linked proteins composed of monomeric (25%), dimeric (21%), trimeric (18%), tetrameric (15%), pentameric (10%), and hexameric (11%) species (Figure 1, lane 2). Following dialysis, heating the formaldehyde-fixed sample in 20 mM Tris-HCl with 2% SDS at pH 4 resulted in an almost 4-fold increase in monomeric protein (Figure 1, lane 3) [4]. To mimic the ethanoldehydration step typically performed during histological processing of FFPE tissues, the formaldehyde-fixed RNase A was precipitated and incubated in 100% ethanol for 1 hr, 24 hr, or 1 week. SDS-PAGE revealed that the formaldehyde-fixed, ethanol-treated samples were as highly cross-linked as the original formaldehyde-fixed samples (Figure 1, lanes 4, 6, and 8). However, after heating in the Tris-SDS recovery buffer at 100°C for 20 min, followed by 60°C for 2 hr, no reversal of the formaldehyde-induced cross-links was observed (Figure 1, lanes 5, 7, and 9) [4].
Figure 1:SDS-PAGE of formaldehyde-fixed RNase A before and after protein retrieval. Lane M: molecular weight marker; lane 1: native RNase A; lane 2: formaldehyde-fixed RNase A after the removal of excess formaldehyde; lane 3: formaldehyde-fixed RNase A sample from lane 2 after retrieval in 20 mM Tris HCl, pH 4.0, with 2% SDS; lanes 4, 6, and 8: formaldehyde-fixed RNase A after incubation in 100% ethanol for 1 hr, 24 hr or 1 week, respectively; lanes 5, 7, and 9; 1 hr, 24 hr or 1week formaldehydefixed, ethanol-treated RNase A after retrieval in 20 mM Tris HCl, pH 4.0, with 2% SDS. The RNase A samples were heated at 100°C for 20 min, followed by 60°C for 2 hr.
The above results suggested that the inability to recover monomeric protein by heat treatment following the incubation of formaldehydetreated proteins in ethanol may result from a combination of cross-link formation and a change in protein conformation. Thus, we examined the structural properties of RNase A treated with formaldehyde and ethanol using circular dichroism (CD) spectroscopy. The far- UV spectrum is sensitive to the secondary structure of the protein. Fixation in 10% formalin for up to 1 week did not significantly alter the secondary structure of RNase A (Figure 2A, profile 2) relative to the untreated protein (Figure 2A, profile 1). Additionally, native, unfixed RNase incubated in 100% ethanol for 1 week rapidly reverted back to its native structure after the ethanol was removed (Figure 2A, profile3). However, when the formaldehyde-fixed RNase A was precipitated under ethanol, there was a significant decrease in band intensity, even after the ethanol was removed and the pellet was resuspended in PBS. The profile changed to one with a single negative peak around 215 nm, characteristic of a structural transition from an α+β to an all-β protein conformation (Figure 2A, profile 4). The near UV-spectra (Figure 2B) also revealed that the tertiary structure of the formaldehydefixed RNase A became significantly disordered after incubation under ethanol [4]. These structural changes are analogous to those seen in proteins that form amyloid fibrils.
Figure 2:The effect of ethanol on protein structure: Far-UV (A) and Near-UV (B) CD spectra of 0.65 mg/ml solutions of RNase A. Profile 1: native RNAse A; profile 2: native RNase A incubated under 100% ethanol for 1 week and then rehydrated in phosphate buffer; profile 3: RNase A kept in 10% formaldehyde for 1 week; profile 4: RNase A fixed in 10% formaldehyde, incubated under 100% ethanol for 1 week, and then rehydrated in phosphate buffer.
How elevated pressure facilitates reversal of formaldehydeinduced protein aggregates
To summarize, the above studies strongly suggest that formaldehydetreated proteins can aggregate and adopt a β-sheet secondary structure at ethanol concentrations >90% [19]. In this form, the regions of the protein inducing its aggregation are those rich in hydrophobic amino acids, which form β-sheet structures stabilized by both intermolecular hydrogen bonds and formaldehyde cross-links [20]. This observation led us to hypothesize that a major obstacle to the reversal of protein formaldehyde modifications in FFPE tissues is the inability to fully re-hydrate these strongly associated protein aggregates by heating alone. We further reasoned that supplementing heat treatment with high hydrostatic pressure would facilitate the re-hydration of such protein aggregates and promote the reversal of formaldehydeinduced protein modifications. Under elevated hydrostatic pressure, cavities in proteins become filled with water molecules, which lead to the hydration of the interior of the protein [21,22]. Hydration of the buried hydrophobic residues induces protein unfolding because the change in molar volume associated with unfolding is negative and thus energetically favored at elevated pressure [23]. Further, formaldehydeinduced protein modifications increase protein stability [24] and can increase the thermal denaturation temperature of fixed proteins to >100°C [18]. Because the change in molar volume associated with unfolding is negative, the thermal transition temperature decreases with increasing pressure [22,25], thus counteracting the stabilizing effect of formaldehyde cross-links. Accordingly, there is a sound thermodynamic basis for believing that increased hydrostatic pressure, along with heat, will facilitate the recovery of proteins from FFPE tissues and promote the reversal of protein-formaldehyde modifications.
Elevated pressure improves recovery of proteins from FFPE tissue surrogates and their analysis by mass spectrometry
To further study the effects of ethanol dehydration and paraffin embedding on proteins, we developed a model system called a “tissue surrogate”, which consists of one or more proteins that form a gellike plug when treated with formaldehyde at protein concentrations exceeding 75 mg/ml. These tissue surrogates have sufficient physical integrity to be processed using routine histological methods. A variety of published extraction buffers and antigen retrieval-based heating protocols were examined for their ability to recover proteins from tissue surrogates [10]. Protein recovery using heat alone was generally modest and studies with multi-protein tissue surrogates revealed extraction bias, meaning that the composition of the solubilized proteins did not match that of the corresponding tissue surrogate [10].
As discussed above, our studies indicated using heat alone was insufficient to recover high quality protein extracts from FFPE tissue. Elevated pressure promotes water penetration into the inner core of proteins, causing denaturation, whereas heat alone causes protein unfolding followed by aggregation [26]. Consequently, we hypothesized that the combined effects of heat and elevated pressure would facilitate the re-hydration of highly aggregated proteins in the tissue surrogate, greatly improving protein solubilization while simultaneously reversing protein-formaldehyde adducts and cross-links.
We initially utilized a battery of extraction conditions first used to enhance immunohistochemistry on FFPE tissue, including heating at 80-100°C, and in a pressure cooker, which utilizes pressures of 1.15 times atmospheric pressure (approximately 17 psi) to superheat water to 120°C [27]. In initial studies, proteins extracted from a lysozyme tissue surrogate at 80-120°C remained highly cross-linked, with total protein content consisting of ~20% monomeric protein and 80% multimeric protein by SDS-PAGE. However, when the lysozyme surrogate suspension was heated at 80°C at pH 4 for 2 hr under 40- 45,000 psi of pressure (3000 times atmospheric pressure), 100% of the protein was recovered in the soluble phase, and almost complete reversal of the formaldehyde-induced protein adducts and crosslinks was observed (data not shown) [10,11]. Initial elevated pressure extraction experiments were carried out using a home built pressure instrument. Samples were heated at 65–100°C under a constant pressure of 45,000 psi in stainless steel reaction vessel coupled to a manually operated HiP High Pressure Generator (High Pressure Equipment Company, Erie, PA, USA). This instrument uses a hydraulic screw pump to compress fluid in order to generate pressure. Since water is virtually non-compressible, in the event of a system failure, discharge forces are minimal and do not present a hazard to equipment or personnel [11]. A NEP 2320 Barocycler (Pressure Biosciences) modified by the manufacturer to hold isobaric pressure was also used to develop pressure-assisted extraction protocols [13]. Pressure cycling technology, consisting of repeated cycles of high pressure to low pressure, has been used to recover proteins, lipids and nucleic acids from fresh tissue [28]. Our laboratory group investigated using pressure cycling to augment protein recovery from FFPE tissue surrogates, but determined that static pressure and temperature over 65°C were more effective in reversing formaldehyde cross-links (unpublished data).
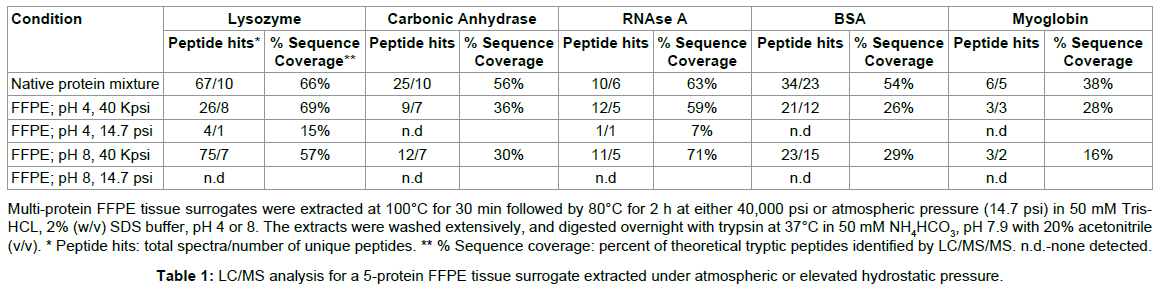
To better mimic the complex mixture of proteins in tissue, we constructed an FFPE tissue surrogate consisting of five proteins with varying abundances, molecular weights, isoelectric points, and secondary structures: lysozyme, carbonic anhydrase, ribonuclease A, BSA, and myoglobin (55:15:15:10:5 w/w). The addition of high hydrostatic pressure (40,000 psi) to augment heat treatment (100°C for 30 min, followed by 80°C for 2 hr) dramatically improved protein extraction efficiency from this multi-protein FFPE tissue surrogate, from ~25% to 96%. By SDS-PAGE, the high-pressure extracted tissue surrogate samples (Figure 3, lanes 2 and 3) consisted of a number of well-resolved bands with the same mobility as the unfixed component proteins (Figure 3, lane 1). In contrast, only lower molecular species were extracted at low (atmospheric) pressure (Figure 3, lane 4) [12].
Figure 3:Elevated pressure improves protein extraction from model FFPE tissue surrogates. FFPE tissue surrogates were heated in 50 mM Tris, pH 8+2% SDS at either elevated pressure (40,000 psi) or atmospheric pressure. The electrophoretic mobility of the tissue surrogate extracts were compared to the native, unfixed tissue surrogate mixture by 1D-PAGE. Lane M: molecular weight marker; lane 1: native, unfixed tissue surrogate mixture; lane 2: FFPE tissue surrogate with 2.5% myoglobin after retrieval at 40,000 psi; lane 3: FFPE tissue surrogate with 5% myoglobin after retrieval at 40,000 psi,; lane 4: FFPE tissue surrogate with 5% myoglobin after retrieval at atmospheric pressure (14.7 psi).
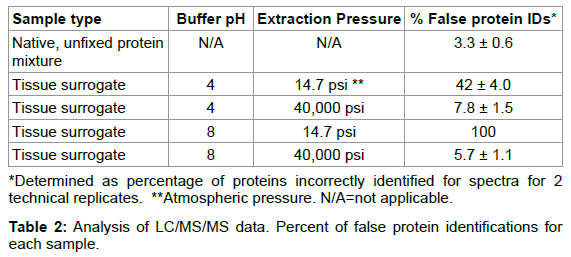
Because the multi-protein tissue surrogate has a defined protein composition, we employed this system for our quality evaluation by LC/MS. We found that the addition of elevated pressure to a wellestablished heat-induced protein extraction protocol [1] improved the proteomic analysis of the FFPE tissue surrogate. For example, the LC/MS trace of the tryptic digests of the surrogates extracted at atmospheric pressure with heat at pH 4 or 8 showed a number of broad, late eluting peaks, suggesting that the material was either poorly digested, or remained cross-linked (Figure 4, panel C, pH 8). There were only a total of 5 correctly identified spectra, representing 2 unique peptides (false ID rate of 42%), for the surrogate extracted at pH 4 and ambient pressure, and no correctly identified tryptic peptides for the surrogate extracted at pH 8 and ambient pressure (Tables 1 and 2) [12]. In contrast, the surrogates extracted with heat and elevated pressure (Figure 4, panel B) compared favorably with the corresponding native, unfixed protein mixture (Figure 4, panel A). The tryptic digest for both the unfixed protein mixture and pressure-extracted samples eluted between 10 and 40% acetonitrile with no late-eluting peaks. The sequence coverage map (percent of theoretical tryptic peptides identified for each component protein) suggested that essentially unmodified proteins were retrieved from the pressure-extracted FFPE tissue surrogates. BSA, which is known to form cross-links with lysozyme in solution [29], was identified with 29% (pH 8) and 26% (pH 4) sequence coverage when extracted from the multi-protein FFPE tissue surrogate at 40,000 psi (Table 2). RNase A and lysozyme, which have a high number of formaldehyde-reactive residues, were identified with sequence coverage comparable to the native protein mixture (59 and 69% sequence coverage, respectively) at pH 4 at 40,000 psi. Myoglobin, which was included as a low-abundance component, was identified by 2 or more fully tryptic peptides in the pressure-extracted multi-protein FFPE tissue surrogates [12]. Thus, we demonstrated that heat augmented with high pressure improved the total recovery of protein and eliminated the extraction bias seen at atmospheric pressure.
Figure 4:Quality comparison of MS profiles of native protein mixture and tissue surrogate extracts. FFPE tissue surrogates were heated in 50 mM Tris, pH 8+2% SDS at either elevated pressure (40,000 psi) or atmospheric pressure. The extracts were analyzed by LC/MS and the MS traces of each tissue surrogate extract were compared to the native, unfixed protein mixture. A) native, unfixed tissue surrogate mixture; B) FFPE tissue surrogate retrieved at 40,000 psi; C) FFPE tissue surrogate retrieved at atmospheric pressure (14.7 psi).
To determine if pressure was accelerating formaldehyde adduct reversal, soluble solutions of formalin-fixed RNase A were heated at 55°C or 65°C for 3.5 hr at 14.7-40,000 psi so that the rate of intermolecular cross-link reversal could be studied independent of protein solubilization, which would not be possible using tissue surrogates (Figure 5) [12]. At 55°C, the percent of monomeric protein was constant, with approximately 82% of the RNase migrating as crosslinked oligomers and 18% of protein migrating as monomeric protein as measured by SDS-PAGE. When the fixed RNase A solutions were incubated at 1 atmosphere and 65°C, the majority of the intermolecular cross-links were reversed, with 62% of protein migrating as monomeric protein by SDS-PAGE. However, at pressures between 5,000-40,000 psi, the amount of monomeric protein decreased to 40-36% of the total protein. These results suggest that the application of elevated pressure does not enhance protein recovery from FFPE tissue by accelerating the rate of formaldehyde adduct reversal [12]. Instead, the reaction rate was modestly decreased by pressure, which may be explained by other studies in which elevated pressure has been shown to protect proteins from thermal denaturation [30] and to inhibit other chemical reactions, such as the Maillard reaction between glucose and lysine [31].
Figure 5:Effect of elevated pressure on the rate of cross-link reversal; percentage of monomeric protein recovered. 1 mg/ml solutions of RNase A was incubated in 10% phosphate buffered formalin for one hour, and the excess formaldehyde solution was exchanged for 1 X TAE buffer, pH 4. The aqueous fixed RNase A solution consisted of 18 percent monomeric and 82 percent multimeric protein by 1-D SDS-PAGE. Aliquots of the formalin fixed solution were incubated at 14.7–40,000 psi for 3.5 hr at either 55°C (squares) or 65°C (triangles). The heat-treated samples were separated by SDS-PAGE and the gel bands were integrated to determine the percentage of monomeric protein at each pressure.
To investigate the effect of pressure on protein solubilization, we used a lysozyme tissue surrogate to examine the effects of elevated pressure on average protein aggregate size. The average particle size, as measured by dynamic light scattering, of samples extracted at atmospheric pressure was 200 ± 55 nm, suggesting that the solubilized fraction remained highly cross-linked, which was confirmed by SDSPAGE (not shown). There was a rapid decrease in particle size with increasing hydrostatic pressure, with a measured average of 40-50 nm after 10,000 psi (Figure 6). Recovery of monomeric protein, as shown by SDS-PAGE, indicated that the decrease in particle size corresponded to the reversal of formaldehyde-induced protein cross-links. These results indicate that pressures above 10,000 psi are necessary for optimal recovery of proteins from FFPE tissue. These results suggest that elevated hydrostatic pressures improves the recovery of proteins from FFPE tissue surrogates by hydrating and promoting solubilization of the protein aggregates, allowing for the subsequent reversal (by hydrolysis) of formaldehyde-induced protein adducts and cross-links [12].
Figure 6: Effect of elevated pressure on aggregate size. Lysozyme tissue surrogates were incubated in 50 mM Tris-HCl buffer, pH 4 with 2% SDS and 0.1 M glycine at 100°C for 2 hr at pressures ranging from atmospheric pressure (14.7 psi) to 50,000 psi. The average particle size of the solubilized protein was measured by dynamic light scattering to determine the degree of protein aggregation.
Elevated pressure improves the proteomic analysis of FFPE liver tissue
Encouraged by our initial physical and mechanistic studies, we applied the pressure-assisted extraction conditions to biologically relevant FFPE tissues, hoping to reduce extraction bias, better recover intact proteins, and to yield tryptic digests that more closely resembled those from matched frozen tissue [13]. FFPE mouse liver, 30 d or 1 y following tissue processing, was extracted using one of two buffers commonly used for proteomic studies of FFPE tissues [1,2]. For both protocols, individual 10 μm sections of FFPE mouse liver were cleared of paraffin with xylene, and rehydrated through a series of graded alcohols (100%, 85% 70%) and water as previously described [11]. For the first extraction trial, whole tissue sections from the 30 d old FFPE liver were homogenized in 50 mM Tris-HCl, pH 7, 2% SDS [1], which is referred to as extraction buffer one (EB1). Half of the tissue homogenate was heated at 100°C for 30 min followed by 80°C for 2 hr at 40,000 psi, while the remaining homogenate was extracted using the same heating protocol, but at atmospheric pressure (14.7 psi, heat alone). This was compared to matched fresh frozen liver extracted in the same buffer, but on ice at atmospheric pressure. Approximately ~80% of the total protein was solubilized by heating at elevated pressure (40,000 psi) in EB1 buffer, relative to fresh-frozen liver from the same animal (n=3). This represents a 4-fold increase over FFPE tissue extracted with heat alone. Similar improvement in total protein recovery was seen with the same FFPE tissue block after storage for 1 y when extracted with 100 mM Tris-HCl, pH 8, 100 mM dithiothreitol (DTT), 4% SDS, which is referred to as extraction buffer two (EB2) [2], for 1 hr at 95°C, with or without elevated pressure. The FFPE liver extracted in EB1 with high pressure and heat exhibited a number of well resolved high and low molecular weight protein bands by SDS-PAGE, corresponding to ~87% of those seen for fresh-frozen liver (Figure 7, lanes 2 and 1, respectively). The FFPE samples extracted with heat alone contained relatively few well-resolved protein bands equivalent to ~25% of those seen in frozen mouse liver (Figure 7, lane 3) [13].
The 30 d old FFPE mouse liver extracted with EB1 using elevated heat and pressure was separated by 1D-PAGE, and each gel lane was excised, digested with trypsin, desalted and analyzed by LC-MS/MS. The corresponding fresh-frozen liver tissue from the same animal was also prepared for LC-MS/MS. The total unique peptide and protein identifications for each tissue type and extraction condition are shown in Table 3 [13]. FFPE tissue extracted with heat alone resulted in the identification of only 5565 unique peptides and 3449 unique proteins. The addition of elevated hydrostatic pressure significantly improved the number of protein identifications, with a total of 9621 unique peptides and 5192 unique proteins. The number of proteins identified from the high pressure-extracted sample was comparable to the number of unique proteins identified from fresh-frozen tissue, which ranged from 4932 for tissue extracted on ice to 4451 for frozen tissue extracted with heat (Table 3). The MS results for the frozen tissue and FFPE mouse liver extracted with elevated pressure in EB1 were searched using GOMiner, a gene ontology program, and the identified proteins were categorized by their sub-cellular compartment and their biological function. The percentages of nuclear, membrane, intracellular and extracellular proteins identified in fresh and FFPE liver were virtually identical as were the results for classification by biological function (not shown) [13].
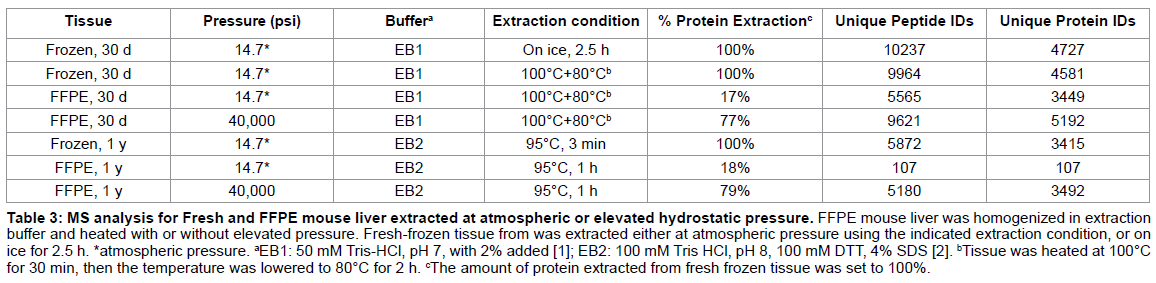
To address the effect of long-term storage of the FFPE specimens, the mouse liver samples were investigated after an additional 11 months of storage (1 y-old sample). To show that extraction efficiency could be improved by the addition of pressure, regardless of buffer, both the long term stored frozen and FFPE mouse liver samples were extracted using a modification of a heat-induced extraction protocol recently published by Ostasiewicz et al. [2]. Specifically, individual 10 μm sections of the FFPE liver were cleared of paraffin and homogenized in EB2 without any microdissection. The homogenates were extracted in a heating block with constant shaking at 95°C for 1 hr, or at 95°C for 1 hr at 40,000 psi. Both samples were separated by1-D SDS-PAGE or 2-D gel electrophoresis. We were able to identify 3492 non-redundant proteins in the 1 year old FFPE liver extracted at 40,000 psi and 95°C, which was comparable to the matched fresh-frozen mouse liver (Table 3) [13].
Our studies show that the addition of elevated pressure to hightemperature FFPE extraction methods affords significantly improved proteome coverage for archival tissue. An increase in pressure to 40,000 psi, to augment heat treatment, improved the extraction efficiency of intact proteins by approximately 4-fold for FFPE mouse liver tissue, and increased the number of unique proteins identified by up to 30-fold in FFPE tissue stored for 1 year [13]. In another recent study, Fu et al. [32] reported a 13% increase in proteins identified from 15 year old archival human aorta extracted with elevate pressure. These results indicate that elevated pressure-assisted extraction is a promising approach to improving extraction of proteins from FFPE tissue for proteomic analysis. In addition, the instrumentation is not out of reach for the average laboratory. A home-built instrument can be constructed from reaction vessels and a manually operated high pressure piston screw pump commercially available from High Pressure Equipment Company (Erie, PA) for less than $5000. The construction and operation of this pressure system has been described in detail previously [11]. A turn-key pressure cycling instrument is also available from Pressure Biosciences, (South Easton, MA), and has been employed for the extraction of archival, formalin-fixed tissue [13,32].
As proteomic and MS technologies continue to mature it is imperative that sample preparation methods do likewise. While extracting proteins from FFPE tissues in the form of tryptic peptides has been sufficient to this point [33], it ultimately limits the types of proteomic analyses that can be done. As top-down MS sequencing technology and the use of reverse-phase microarrays becomes more common, the ability to extract and analyze intact, high quality proteins from FFPE tissue will become more important. Top-down sequencing facilitates the measurement of combinations of modifications, such as phosphorylation, and the direct quantitation of specific protein isoforms and splice variants. None of these measurements is directly obtainable using approaches in which proteins are digested into peptides. As additional data concerning disease-specific biomarker becomes available, aberrant protein modifications that cause diseases such as cancer will continue to be discovered. By extracting intact proteins from the seemingly inexhaustible source of FFPE tissues, the diagnostic or prognostic efficacy of these discoveries could potentially be validated using orthogonal methods such as Western blotting, immunohistochemistry, immunoassays [34], and structural and interaction proteomics.
The authors have been funded, in part, with federal funds under grant 1R21 CA134359 from the NCI Innovative Molecular Analysis Technologies (IMAT) Program, and the Veterans Health Administration under a Merit Review award. The content of this publication does not necessarily reflect the views or policies of the Veterans Health Administration nor does the mention of trade names, commercial products or organization(s) imply endorsement by the US Government. The authors are named as co-inventors on patents US 8,288,122 B2 and US 8,481,283 B2, ‘pressure-assisted molecular recovery (PAMR) of biomolecules, pressureassisted antigen retrieval (PAAR) and pressure assisted tissue histology (PATH)’.