Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2020)Volume 13, Issue 4
Drought stress is a major abiotic factor causing yield loss in safflower and limited studies have been carried out to dissect the molecular mechanism at EST-SSR level. In this study, a possible relationship between simple sequence repeats (SSRs) distribution and drought stress was studied using three EST libraries of safflower, Ct-D-EST, Ct-NEST and Co-EST. The Ct-D-EST was generated from drought tolerant safflower cultivar A-1; Ct-N-EST and Co-EST were the EST datasets of safflower and its wild progenitor C. oxycanthus, respectively. In total, 156 (45%), 1194 (5%) and 1550 (9%) EST-SSRs were mined from Ct-D-EST, Ct-N-EST and Co-EST libraries, respectively. Comparison of EST-SSRs from Ct-D-EST with that of SSRs from other two libraries showed reasonable differences for each class of repeats. Large variations were observed for dinucleotide repeats in all the libraries. In drought EST-SSRs, only one kind of amino acid was produced by repeat (ATG)14 which encodes met indicating the loci and repeat observed to be 100%. Since three ESTs with SSRs from Ct-D-EST, annotated to putative candidate genes, S-adenosylmethionine synthetase, one-helix protein and myo-inositol 1-phosphate synthetase, did not express with SSRs in the Ct-N-EST, these can be considered as potential candidate genes for drought tolerance. A trinuceotinde SSR, encoding met, was also found in the EST annotated to putative s-adenosylmethionine synthetase, hence, both can be considered as indices for drought tolerance. The genomic resources and the information generated in this study may be useful for the safflower breeders particularly for the development of drought tolerant cultivar.
Drought stress; Safflower; Subtractive hybridization; Simple sequence repeats; Candidate genes
In India, safflower (Carthamus tinctorious L.) is cultivated mainly for the high-quality edible oil extracted from the seeds. valued. It is also grown for the orange-red dye (carthamin) extracted from its flowers used in food and textile industries [1]. Safflower oil is rich in polyunsaturated fatty acids (linoleic acid, 78%) and is nutritionally and medicinally valued. Among the major oilseed crops, safflower is relatively more adaptable to drought and saline conditions [2] however, with reduced crop yield. Wild safflower (Carthamus oxyacanthus) also show high drought tolerance making it is as a suitable source for transferring drought tolerant genes to cultivated species [3,4]. There has been only limited information on genetic control of abiotic stress tolerance in safflower [5-7]. ESTs are short piece of cDNA sequences generated by large-scale single pass sequencing of cDNA clones. They serve as low-cost alternative to whole genome sequencing; they are efficient in gene discovery and development of DNA markers. SSRs (single sequence repeats) are sequences in which one or few bases are tandemly repeated, ranging from 1-6 base pair (bp) long units. They are ubiquitous in eukaryotic and prokaryotic genomes, found in both coding and non-coding regions [8,9]. These features make them powerful genetic markers for a variety of applications ranging from the determination of parentage, genetic relationships and genetic mapping [10,11]. Moreover, identification of drought linked ESTSSRs would immensely benefit plant breeders for screening and selection of genotypes and transfer of specific traits for particular stress tolerance. In recent years, significant efforts have been made to develop the SSR markers in safflower [12-16] but still the number is less compared to the other oilseeds crops like sunflower, soybean, brassica, groundnut etc. In this study, we developed SSRs from three EST collections, drought responsive EST (Ct-D-EST) and ESTs datasets of safflower (Ct-N-EST) and wild safflower (Co- EST). By comparing and analyzing drought SSRs with normal ones, we tried to find a possible relationship between SSRs distribution and drought stress.
Sequence resources and EST assembly
A total of 591 ESTs from our drought subtractive cDNA library generated from safflower cultivar A1 [17] were used and named as Ct-D-EST library. EST dataset with 24,299 sequences of safflower was downloaded from the TIGR Plant Transcript Assemblies database (http://plantta.tigr.org) used as normal ESTs, named as Ct-N-EST library. EST data set 27,255 sequences of wild safflower was downloaded from Compositae genome project (http://compgenomics.ucdavis.edu/) and named as Co-EST library. AllEST sequences were assembled by CAP3 software (http://genome.cs.mtu.edu/sas.html). The non-redundant unigene sequences from drought ESTs, normal ESTs and wild species were used to identify microsatellites.
Identification and comparison of EST-SSR and amino acid distribution with trinucleotide repeats between the Ct-DEST, Ct-N-EST and Co-EST libraries
The unigene sets of all the libraries were searched for SSRs using SSRLocator [18-21] and the abundance of amino acids predicted from the trinucleotide repeats were compared among all the libraries. SSR locator was used to search SSRs (motifs ranging from 1 to 10 nucleotides) in order to evaluate the pattern of EST-SSR distribution among all libraries. The repeat number parameters were as follows: ≥ 20 for mono, ≥ 10 di, ≥ 7 for tri, ≥ 5 for tetra, ≥ 4 for penta and hexa and ≥ 3 for hepta, octa, nona and deca nucleotide, respectively. The space between imperfect SSRs were kept at 5.
Primer designing and predicting the amino acid composition
For each microsatellite-containing EST, primers were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) by running the software in a batch mode with the assistance of the SSR locator interface module. The major parameters for primer design were as follows: PCR product size 100–300 bp, primer length 15–25 bp with 20 bp as the optimum, optimum annealing temperature 55– 61°C with 60°C as the optimum, and a minimum GC content of 30%, with 50% being the optimum. After predicting the amino acid composition of ESTs containing trinucleotide tandem repeats, the number of amino acid loci and number of amino acid repeats were compared among the libraries.
Assessment of functional relevance of unigenes having SSRs
Unigene sequences from Ct-D-EST and Ct-N-EST libraries containing microsatellites were used for similarity search using Blast2GO [22] to identify their putative function and were run against the non-redundant (nr) protein database of the NCBI (http://www.ncbi.nlm.nih.gov/blast); the obtained hits were compiled [23]. Unigene sequences not showing any match were considered as unique to the safflower species.
Frequency and distribution of EST-SSRs in Ct-D-EST, Ct- N-EST and Co-EST libraries
In plants, microsatellites derived from EST sequences (EST-SSRs) have been proposed to be better candidates for gene tagging and are preferred over genomic-SSR markers for plant improvement programs owing to their higher interspecific transferability rate [24]. Microsatellites are the current method of choice because they can be traced either in protein-coding or non-coding regions with high mutability [25]. EST-SSRs by following the behavior of SSRs in the coding part of genome also provide valuable information about the effects of SSRs on drought stress mechanism and functional alteration of genes during the drought stress. Total, 52145 ESTs were analyzed by SSR Locator; 591 ESTs belonged to Ct-D-EST, and 24,299 ESTs belonged to Ct-N-EST and 27,255 Co-EST (Table 1). After assembly, unigenes were identified and the average lengths of the EST sequences in Ct-D-EST, Ct-N-EST and Co-EST were 494, 754 and 633 bp, respectively. The distribution and occurrence of type of SSR motif repeats in all the three ESTs libraries are presented in Figure 1. The EST-SSRs developed from Ct-D-EST, Ct-N-EST and Co-EST libraries were named as drought SSRs, normal SSRs and wild SSRs, respectively. In Ct-D-EST, the percentages of mono, di, tri, tetra, and penta nucleotide were 92.30%, 5.12%, 0.64%, 1.28% and 0.64%, respectively (Table 2) which shows differential distribution of repeat motif under drought stress. Since only mono, di, tri, tetra, and penta nucleotide repeats were found in Ct-D-EST, the repeats of this level were compared among the libraries. There are reports showing the differential distribution of SSR repeats in crop plants under stress condition. In chickpea, EST SSRs ranging from mono-hexa nucleotide repeats were developed from drought responsive ESTS developed from drought responsive ESTs [26,27]. The percentage of trinucleotide repeats was less in the Ct-D-EST library compared to Ct-N-EST and Co-EST libraries (0.64% versus 38.35; 36.25%, respectively). Comparative analysis of EST-SSR types between Ct-D-EST, Ct-N-EST and Co-EST libraries
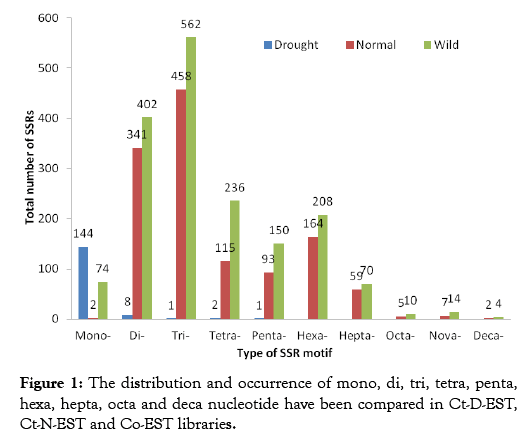
Figure 1: The distribution and occurrence of mono, di, tri, tetra, penta, hexa, hepta, octa and deca nucleotide have been compared in Ct-D-EST, Ct-N-EST and Co-EST libraries.
| Variables | Drought ESTs | Normal ESTs | Wild ESTs |
|---|---|---|---|
| Sequences assembled | 591 | - | 27255 |
| Contings | 53 | - | 3559 |
| Unigenes | 350 | 24299 | 16766 |
| Total SSRs | 156 (45 %) | 1194 (5%) | 1550 (9%) |
| ESTs with one SSR loci | 145 (92%) | 1144 (96%) | 1442 (93%) |
| ESTs with two SSR loci | 11 (8%) | 48 (4%) | 62 (4%) |
| ESTs with three SSR loci | 0 | 2 (1%) | 28 (2%) |
| ESTs with four SSR loci | 0 | 0 | 10 (1%) |
| ESTs with five SSR loci | 0 | 0 | 8 (1%) |
Table 1: Summary of the ESTs assembly and EST-SSRs distribution in Ct-D-EST, Ct-N-EST and Co-EST libraries.
| SSR Repeats | Drought SSRs | Normal SSRs | Wild SSRs |
|---|---|---|---|
| Mononucleotide | 144 (92.30%) | 2 (0.16%) | 74 (4.77%) |
| Dinucleotide | 8 (5.12%) | 341 (28.55%) | 402 (25.93%) |
| Trinucleotide | 1 (0.64 %) | 458 (38.35%) | 562 (36.25%) |
| Tetraucleotide | 2 (1.28%) | 115 (9.63%) | 236 (15.22 % ) |
| Pentanucleotide | 1 (0.64%) | 93 (7.78%) | 150 (9.67%) |
| Friedman test: Chi-Square = 10.8 ; P = 0.005 | |||
| Wilcoxon signed rank test | |||
| Drought-Normal | Z= -2.49 ; P = 0.013 | ||
| Drought-wild | Z= -1.79; p= 0.07 | ||
| Normal -Wild | Z= -1.1; p=0.272 | ||
Table 2: Comparison of expressed SSR on percent basis, by Friedman test and pair wise comparison for each class of SSRs between Ct-D-EST, Ct-N-EST and Co-EST libraries.
Comparative analysis of EST-SSR types from all the libraries gave an insight on the expressed SSR sequences in each class of repeat units. Comparison of 156 SSRs from Ct-D-EST with the SSRs from Ct-N-EST and Co-EST showed large variation in frequency of these repeat motifs. The types of dinucleotide sequences in all the libraries are presented in Figure 2. Five dinucleotides, AT/ AT, GA/TC, TA/TA, TG/CA, and CT/AG were found in all the libraries, while TG/CA and CT/AG were solely detected in Ct-DEST. It is important to note here, the distribution of dinucleotide sequences was not similar among the libraries. The occurrence of the AT/AT was observed to be most variable among Ct-D-EST, CT-N-EST and Co-EST libraries i.e. 22.22%, 0.30% and 7.00%, respectively. The number of normal SSRs (1194) and wild SSRs (1550) were nearly 8 and 11 times, respectively, more than the drought SSRs (156). The sole detection of TG/CA and CT/AG repeats in drought SSRs suggests that a small number of ESTs are sufficient to study the repeat variations in drought stress at ESTSSR level. These results suggest that it would be reasonable to generate a small number of ESTs, in order to develop new ESTSSR marker in crops like safflower where the genomic resources are limited. Similar type of observations were recorded by Torben et al. [28] while comparing the frequency, type and distribution of EST-SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon and Oryza sativa. In the Ct-D-EST library, the distribution of different sequences of expressed tri, tetra and penta nucleotide SSRs were observed hence, we have compared the frequency distribution pattern of these repeats with repeats from other ESTs libraries (Table 3). The repeats ATG, CTTT, AATA and GAAGT occurred only once in Ct-D-EST which contributed 0.65% each. In other plant species, the most frequent trinucleotide repeat motifs were AAC/TTG in wheat, AAG/TTC in soybean, and CCG/GGC in barley, rice, maize and sorghum [29,30]. The previous studies on Arabidopsis and soybean reported abundance of trinucleotide motif AAG [31]. In the case of trinucleotide repeats, only one type of triplet repeat i.e., ATGwas found in Ct-D-EST whereas, 45 and 51 sequence-types, respectively, were detected in normal and wild ESTs (Supplementary Information I). A differential distribution of trinucleotide repeats was highly noticeable in all the libraries. The distribution of triplet repeat ATG was uniform in all the libraries but the percent distribution among the libraries was varied greatly (100%, 2.17% and 4.25%, respectively). It was clear that Co-EST library was rich source of trinucleotide repeats with highest frequency of 565 compared to Ct-N-EST (460) and Ct-D-EST (1). When compared to the total number of repeats to the frequency of occurrence of repeats in Ct-D-EST library, it was interesting to note that the repeats ATG, CTTT, AATA, GAAGT occurred only once. Hence, notable distribution of different sequences of expressed tri, tetra and penta nucleotide SSRs were observed in the present study.
| EST library | Trinucleotide | Frequency | % |
|---|---|---|---|
| Drought | ATG | 1 | 100 |
| Normal | ATG | 10 | 2.17 |
| Wild | ATG | 24 | 4.25 |
| Tetranucleotide | |||
| Drought | AATA | 1 | 50 |
| Normal | AATA | 2 | 2.02 |
| Wild | -----NA----- | 0 | 0 |
| Drought | CTTT | 1 | 50 |
| Normal | CTTT | 8 | 8.08 |
| Wild | CTTT | 2 | 0.91 |
| Pentanucleotide | |||
| Drought | GAAGT | 1 | 100 |
| Normal | GAAGT | 1 | 1.12 |
| Wild | GAAGT | 2 | 1.3 |
Table 3: Comparison and distribution of different sequences of expressed tri, tetra and penta nucleotide SSRs in Ct-D-EST, Ct-N-EST and Co-EST libraries.
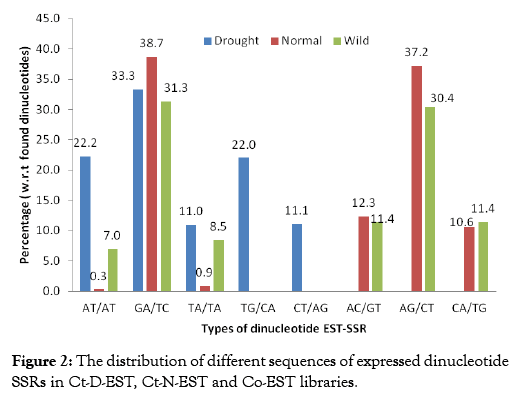
Figure 2: The distribution of different sequences of expressed dinucleotide SSRs in Ct-D-EST, Ct-N-EST and Co-EST libraries.
In -silico PCR results and functional annotation of ESTSSRs
In Ct-D-EST, Ct-N-EST libraries 58 and 916 EST-SSRs had proper flanking regions for primer design which contribute nearly 40% and 77% of the total ESTs. When virtual PCR was run with the SSR Locator software, 33 of the primers (58%) produced suitable fragments in Ct-D-EST and 623 of the primers (48%) produced suitable fragments in Ct-N-EST. The identified primer sets for drought and normal SSRs are presented in Supplementary Information II. To explore the functions of the EST with SSRs, BLASTX was performed against the non-redundant protein (nr) databank of NCBI. The BLASTX hits were classified based on similarity scores, >80%, <80% and ‘not available in the database’. The results showed that 78 sequences (53%) of drought and 131 sequences (11%) for normal ESTs were found to have no matches in the database. A comparative functional annotation of ESTSSRs between Ct-D-EST and Ct-N-EST libraries is presented in Supplementary Information III. In addition, annotated proteins for tri, tetra and penta nucletotide EST-SSRs were compared between the libraries (Supplementary Information IV). Normal ESTs with SSRs were annotated to proteins having distinct molecular functions such as, binding, catalytic, transport, enzyme regulators, structural activities in different biological processes, and cellular and sub-cellular organization. Among the annotated ESTSSRs in Ct-D-EST, nearly 58 were expressed and these proteins did not express with SSRs in the Ct-N-EST library (Supplementary Information III). The EST sequences with dinucleotide repeats did not match to any known protein in BLASTX analysis. In human beings, trinucleotide, tetrameric and even dodecameric repeats were found to have role in causing neurodegenerative disorders and cancers [17,18]. Hence, in our study, more emphasis was given to identify the protein functions of EST-SSRs with tri, tetra and penta nucletotide from Ct-D-EST library. An EST-SSR with trinucleotide repeat (ATG) from Ct-D-EST matched with s-adenosylmethionine synthetase protein and the same repeat was also sole responsible for the production of met. In broomcorn millet, expression pattern of PmSAMS (s-adenosylmethionine synthetase) was observed during drought stress and after re-watering indicated its involvement in drought stress [32]. In plants, drought stress causes the alterations in the chemical composition and physical properties of the cell wall and, such change may involve the genes encoding S-adenosylmethionine synthetase [33]. The EST-SSR containing sequences yielded two tetranucleotide repeats CTTTT out of which one (Sequence CtDI_A_54 containing SSR repeat) did not matched to any known protein, indicating it may be specific to safflower and further studies are required to understand its role in drought tolerance. Another repeat (Sequence ID CtDI_B_22 containing SSR repeat) matched with one-helix protein. Primary function of one-helix protein is to absorb light through chlorophyll [34]. EST-SSR containing pentanucleotide repeat (GAAGT) matched with myo-inositol 1-phosphate synthase protein which was shown to play an important role in drought tolerance in chickpea [35]. Expression of MIPS2 was observed in different seed developmental stages in response to heat stress in rice and Arabidopsis [36-38]. In particular, interference of SSRs with S-adenosylmethionine synthetase, one-helix protein and myoinositol 1-phosphate synthetase proteins play important roles in plant response to drought stress. These genes can be considered as potential candidate genes for drought tolerance in safflower because these genes did not express with SSRs in the Ct-N-EST library. The SSRs, ATG, CTTT, AATA and CTTTT were linked to drought stress in safflower. Similarly, SSRs were identified from drought-responsive ESTs in wheat, soybean, maritime pine and the SSR types were CAG, ATA and AT, respectively. The genes reported in present study are novel as reported by earlier reports and linked to the EST-SSR markers. The SSRs specific to drought ESTs can be used as markers for drought tolerance in safflower.
Overall amino acid distribution in all the ESTs libraries
Translating the EST-SSRs to their corresponding amino acids provided some clues about differences and similarities between drought and normal SSRs at the protein level. The expression pattern of EST-SSRs at the amino acid level was quite different in all the libraries (Table 4). In drought EST-SSRs, only one kind of amino acid was produced by repeat (ATG)14 which encodes met indicating the loci and repeat observed to be 100%. In addition, the type of expressed amino acids was notably different between normal and wild SSRs. Ser was the most abundant amino acid found in normal and wild SSRs which contributed nearly 13.63% and 9.80%, respectively, of the total loci and nearly 14.53% and 11.78, respectively, of the total repeat in both libraries. The maximum loci percentage was recorded for Pro (8.67%), Phe (8.30%) and Leu (8.18%) which were abundant in normal SSRs. EST-SSR containing trinucleotide repeat (ATG) was sole responsible for the production of met and when this sequence was annotated to s-adenosylmethionine synthetase protein. Hence, from our results we interpret that the abundance of met in drought tolerant cultivar A-1 genotype may have a role in drought tolerance. In plant cell it has been reported that methionine as a fundamental metabolite through its first metabolite S-adenosylmethionine synthetase [39]. In contrast to our results, Thippeswamy et al. [20] reported the water stress tolerance mechanism in safflower was controlled by pro which was due to increased activity of P5CS. In safflower, the range of met is 1.4-1.5%, pro 8.5-9.5% and the range of pro is higher than the met. Our study clearly indicated that the increase in the level of met in drought stress condition may have important role in drought tolerance in safflower. The plant protein showing the strongest response to drought stress was identified as methionine synthetase, which is involved in both the de novo synthesis of methionine and in the regeneration of the methyl group S-adenosyl-L-methionine [40]. In plants, it has been estimated that about 20% of the methionine is incorporated into proteins [41]. It is suggested that the s-adenosylmethionine synthetase may have major role along with met for drought tolerance in safflower. Hence, trinucleotide SSR which produces met and the gene s-adenosylmethionine synthetase both can be considered as indices for drought tolerance in safflower (Table 5).
| Repeats | Drought | Normal | Wild | Friedman/Wilcoxon Sign test values |
|---|---|---|---|---|
| Dinucleotides | 40.2 (20.09) | 0.8 (0.78) | 34.1 (36.62) | 13.4 ** |
| Trinucleotides | - | 8.3 (11.15) | 10.22 (11.42) | -1.46NS |
| Tetranucleaotides | - | 1.3 (2.5) | 2.8 (3.12) | -4.19** |
| Pentanucleaotides | - | 0.8 (1.02) | 1.3 (1.240 | -3.69** |
| Figures in parentheses are standard deviations | ||||
Table 4: The distribution of expressed SSRs between all each class of SSRs tandem repeats di, tri, tetra and pent nucleotide between Ct-D-EST, Ct-N-EST and Co-EST libraries by Friedman/Wilcoxon Sign test.
| Drought SSRs | Normal SSRs | Wild SSRs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid | No. of Loci | Total Repeats | %loci | % repeats | No. of Loci | Total Repeats | % loci | %repeats | No. of Loci | Total Repeats | % loci | % repeats |
| Ala | 0 | 0 | 0 | 0 | 8 | 44 | 0.99 | 0.84 | 20 | 92 | 1.96 | 1.39 |
| Arg | 0 | 0 | 0 | 0 | 40 | 242 | 4.96 | 4.62 | 50 | 334 | 4.90 | 5.06 |
| Asn | 0 | 0 | 0 | 0 | 28 | 136 | 3.47 | 2.60 | 22 | 120 | 2.16 | 1.82 |
| Asp | 0 | 0 | 0 | 0 | 35 | 216 | 4.34 | 4.12 | 68 | 466 | 6.67 | 7.06 |
| Cys | 0 | 0 | 0 | 0 | 4 | 25 | 0.50 | 0.48 | 14 | 90 | 1.37 | 1.36 |
| Gln | 0 | 0 | 0 | 0 | 38 | 235 | 4.71 | 4.49 | 20 | 106 | 1.96 | 1.61 |
| Glu | 0 | 0 | 0 | 0 | 56 | 382 | 6.94 | 7.29 | 96 | 592 | 9.41 | 8.96 |
| Gly | 0 | 0 | 0 | 0 | 33 | 179 | 4.09 | 3.42 | 46 | 222 | 4.51 | 3.36 |
| His | 0 | 0 | 0 | 0 | 60 | 388 | 7.43 | 7.41 | 34 | 220 | 3.33 | 3.33 |
| Ile | 0 | 0 | 0 | 0 | 59 | 372 | 7.31 | 7.10 | 84 | 532 | 8.24 | 8.06 |
| Leu | 0 | 0 | 0 | 0 | 66 | 461 | 8.18 | 8.80 | 98 | 666 | 9.61 | 10.08 |
| Lys | 0 | 0 | 0 | 0 | 36 | 245 | 4.46 | 4.68 | 30 | 204 | 2.94 | 3.09 |
| Met | 1 | 14 | 100 | 100 | 13 | 94 | 1.61 | 1.79 | 40 | 262 | 3.92 | 3.97 |
| Phe | 0 | 0 | 0 | 0 | 67 | 481 | 8.30 | 9.18 | 72 | 414 | 7.06 | 6.27 |
| Pro | 0 | 0 | 0 | 0 | 70 | 423 | 8.67 | 8.08 | 32 | 160 | 3.14 | 2.42 |
| Ser | 0 | 0 | 0 | 0 | 110 | 761 | 13.63 | 14.53 | 100 | 778 | 9.80 | 11.78 |
| Term | 0 | 0 | 0 | 0 | 21 | 117 | 2.60 | 2.23 | 56 | 424 | 5.49 | 6.42 |
| Thr | 0 | 0 | 0 | 0 | 36 | 263 | 4.46 | 5.02 | 48 | 320 | 4.71 | 4.85 |
| Trn | 0 | 0 | 0 | 0 | 8 | 46 | 0.99 | 0.88 | 26 | 140 | 2.55 | 2.12 |
| Tyr | 0 | 0 | 0 | 0 | 7 | 42 | 0.87 | 0.80 | 24 | 188 | 2.35 | 2.85 |
| Val | 0 | 0 | 0 | 0 | 12 | 86 | 1.49 | 1.64 | 40 | 274 | 3.92 | 4.15 |
| Total | 1 | 14 | 807 | 5238 | 1020 | 6604 | ||||||
Table 5: The expression pattern of EST-SSRs at the amino acid level between Ct-D-EST, Ct-N-EST and Co-EST libraries at trinucleotide level.
In this study, we developed and compared EST-SSR marker from the Ct-D-EST, Ct-N-EST and Co-EST libraries in safflower. We proved that the distribution, frequency and type of EST-SSRs developed from Ct-D-EST were different than the repeats from Ct-N-EST and Co-EST libraries. It is suggested that further studies are required to understand the role of identified putative candidate genes in drought tolerance of safflower. Identification of putative candidate genes underlying drought stress will facilitate understanding of molecular mechanisms controlling drought tolerance and this would lead to genetic improvement of safflower. The EST-SSR markers developed in silico from our study should be validated in wet lab which may prove important assets for the molecular breeding, screening and development of drought tolerant genotype in safflower.
Authors greatly acknowledge the DBT, New Delhi for financial support to CS in the form of research project.
Citation: Sudhakar C, Thippeswamy M, Sivakumar M, Sudhakarbabu O, Dudhe MY (2019) In silico Mining of EST-SSRs from the Drought Tolerant ESTs in Safflower. J Proteomics Bioinform 12:8. doi: 10.35248/0974-276X.1000506
Received: 21-Oct-2019 Accepted: 20-Dec-2019 Published: 27-Dec-2019 , DOI: 10.35248/0974-276X.19.12.506
Copyright: © 2019 Sudhakar C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.