Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Editorial - (2017) Volume 10, Issue 6
Most of the commonly used indices of codon usage bias are Relative synonymous codon usage (RSCU), Effective Number of Codon (ENC), Codon bias Index (CBI) and Intrinsic codon bias Index (ICDI). The parameters and algorithms for each index are described below:
The relative synonymous codon usage is the ratio of the observed frequency of a codon to the expected frequency of a codon if all the synonymous codons for a particular amino acid are used equally [1]. If the RSCU value of a codon is greater than one that the codon is frequently used than expected whereas if the RSCU value of a codon is less than one that the codon is less frequently used than expected. If the RSCU value of a codon is <0.6, it represents the under-represented while if the RSCU value of a codon >1.6, it represents over-represented. RSCU is calculated as:
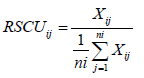
Where, Xij is the frequency of occurrence of the jth codon for ith amino acid (any Xij with a value of zero is arbitrarily assigned a value of 0.5) and ni is the number of codons for the ith amino acid (ith codon family).
ENC is a widely used measure of codon usage bias and its value is dependent upon the nucleotide composition of a gene. Its value ranges from 20 to 61 and higher value means lower codon usage bias and vice-versa. If the value is 20, it signifies only one codon is used for each amino acid and if the value is 61 it denotes all the codons are equally likely to code for the same amino acid. ENC is measured as:
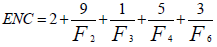
Where Fk (k=2, 3, 4, 6) is the mean of Fk values for the k-fold degenerate amino acids [2].
The codon bias index is also the measure of codon usage bias, based on the degree of preferred codons used in a gene, like to the frequency of optimal codons. Its value ranges from 0 to 1. The 0 indicates random choice of codon whereas 1 signifies only the preferred codons used in a gene while less than zero denotes non-preferred codons used more in the gene [3]. CBI can be calculated as:
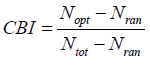
The ICDI is a parameter of codon usage bias and it is not based on optimal codons. The value of ICDI ranges from 0 to 1 where 0 signify for equal usage and 1 for extremely high-biased genes. The ICDI value greater than 0.5 indicates high bias whereas its value lowers than 0.3 means little bias. The ICDI is computed based on Sa values for each of the 18 amino acids with k-fold degeneracy [4].
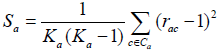
Where rac is the relative synonymous codon usage and ka is the degeneracy of amino acid in the sequence. The value of the index is then computed as:
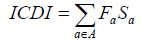
The ICDI gives equal weight to all amino acids included, that is, all values of Fa are 1/18.