Journal of Theoretical & Computational Science
Open Access
ISSN: 2376-130X
ISSN: 2376-130X
Research Article - (2024)Volume 10, Issue 3
The Avermectins, part of the 16-membered macrocyclic lactone family, include Ivermectin, Abamectin, Moxidectin, Milbemycin oxime, Doramectin, Selamectin and Eprinomectin. Recent discoveries regarding the anticancer properties of Avermectins have exhibited anticancer effects across various cell lines. This study aims to employ in silico methodologies to assess the therapeutic potential of Avermectin family compounds against the tubulin protein. Molecular docking analyses were conducted using the tubulin protein via the CB-Dock2 webserver, with visualization performed using PyMol software. Ligands were detached using Notepad++ and SWISS-MODEL was utilized to construct a template for missing amino acid residues. Remarkably, the results demonstrated that some of the Avermectin family compounds exhibited high binding scores compared to the reference anticancer drug, Taxol. Specifically, Ivermectin B1a, Selamectin and Doramectin showed the highest scores of -18.0, -9.1 and -8.9 Kcal/mol, respectively. All Avermectin compounds displayed similar affinity and Ivermectin with each exhibiting greater binding affinity than the reference drug Taxol. This research thus provides avenues for investigating tubulin-targeting compounds, suggesting the potential of Avermectin compounds. Key interactions with β-tubulin residues suggest that Avermectins may stabilize microtubules similar to Taxol, providing a strong basis for further in vitro and in vivo studies on their effectiveness as anticancer agents.
Avermectins; Tubuli; In silico; Molecular docking; Anticancer
Cancer is characterized by the uncontrolled growth and proliferation of cells, making it one of the most lethal diseases globally. Given its significant impact on public health, there is an urgent need to explore innovative methods and therapies to combat this disease. Due to its severe effects, scientists are actively pursuing the identification of novel targets and the development of new drugs for cancer treatment. The search for alternative therapeutic approaches is influenced by increasing for effective anticancer agents capable of addressing the diverse complexities and challenges associated with combating cancer [1,2].
Tubulin's α and β monomers are isotypes with distinct amino acid sequences encoded by separate genes. Alpha/beta heterodimers combine to form microtubules, which are essential for cellular division and growth. Microtubules exhibit an essential trait known as dynamic instability, wherein they are highly dynamic structures composed of dimers continuously integrating into and disengaging from the microtubule within cells. Given microtubules' pivotal role in mitosis, they are significant targets for anticancer therapy, a perspective explored through tubulin binding agents. These compounds can disrupt microtubule dynamics, serving as either destabilizers (such as vinca alkaloids and colchicinoids) or stabilizers (like Taxol) of microtubules [3-5].
Avermectins are a class of pharmaceuticals that are produced naturally as a byproduct of the fermentation of Streptomyces avermitilis, which is an actinomycete that is isolated from the soil [6]. Over the years, considerable interest has been drawn towards fermentation-produced, intricate macrocyclic lactones like Ivermectin, Abamectin, Moxidectin, Milbemycin oxime, Doramectin, Selamectin and Eprinomectin (Figure 1) [7]. These compounds have obtained attention due to their natural bioactivity, notable efficacy and absence of cross-resistance and distinctive mechanisms of action [8].
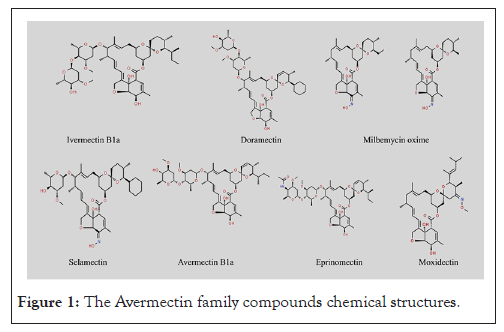
Figure 1: The Avermectin family compounds chemical structures.
Avermectins consist of two classes; major and minor classes. The major classes are the A1a, A2a, B1a and B2a, while the minor classes constitute the A1b, A2b, B1b and B2b. Indeed, Ivermectin, Abamectin, Doramectin, Eprinomectin, Moxidectin and Selamectin represent notable examples of Avermectins [7].
Previous research has suggested that compounds belonging to the Avermectin family possess significant anticancer properties, demonstrating efficacy in impeding the proliferation and progression of tumor cells. Their anticancer potential stems from their ability to interfere with various cellular processes critical for tumor growth and metastasis, including cell cycle regulation, angiogenesis and apoptosis pathways [6, 9-11].
Recent investigations into Ivermectin have revealed its direct interaction with both nematode and human tubulin, even at micromolar concentrations [12]. Experiments have shown that Ivermectin effectively inhibits the proliferation of HeLa cells reversibly, possibly by stabilizing mammalian microtubules similar to Taxol [13]. Additionally, emerging research suggests that Avermectin B1a, possesses anticancer properties by promoting tubulin polymerization akin to Taxol, indicating the potential of Avermectins as strong candidates for anticancer therapy [11].
An advantage of in silico methodologies is their ability to pinpoint novel compounds with favorable characteristics as potential druggable targets prior to synthesis, thereby diminishing the necessity for laborious and costly animal and in vitro experiments [14].In contemporary research, docking studies are significantly influential in exploring the interaction between ligands and proteins and these methodologies are widely employed in various scientific investigations [15].
In this research, we used computational analysis to investigate the molecular interaction between the Avermectin compound and β-tubulin, aiming to determine the molecules displaying the strongest affinity for binding to β-tubulin. We stabilized and confirmed the validity of the modified β-tubulin.
ChemSpider database was used to download the structures of 6 out of the 7 ligands used in this study. These include: Avermectin B1a (Abamectin) molecular formula C48H72O14 (ID: 10286553), Ivermectin B1a (Ivermectin) molecular formula C48H74O14 (ID: 16736314), Moxidectin molecular formula C37H53NO8 (ID: 22901017), Doramectin molecular formula C50H74O14 (ID:8008478), Selamectin molecular formula C43H63NO11 (ID:16738655), Eprinomectin B1a molecular formula and C50H75NO14 (ID: 16736607). Milbemycin oxime was drawn using the Simplified Molecular Input Line Entry System (SMILES). This is because the structure on the ChemSpider database is heavy for the docking purposes.
Protein preparation
The structure of the Tubulin Alpha-Beta Dimer (1TUB) was downloaded from the Protein Data Bank (PDB). Protein was viewed in PyMol and ligands were detected. Notepad++ was used to remove the attached ligands. Detected breaks were also removed using SwissModel. The Fast Adaptive Shrinkage Threshold Algorithm (FASTA) was pasted to model the protein. 88.67% sequence identity was obtained. The modeler protein was downloaded and opened in PyMol to recheck if any existing breaks remained. The downloaded model was in dimer and a particular chain (Chain A) was selected and saved in PDB format for further analysis.
Molecular docking
The docking study of tubulin against the aforementioned Avermectins was performed using an online docking tool known as Cavity-detection guided Blind Docking (CB-Dock). Receptors and ligands are uploaded in the form of PDB files to the docking server separately. The receptor was uploaded with the corresponding ligand at a time. Docking was performed and results were viewed based on Vina score, cavity volume, center, docking size and contact residues.
Molecular docking analysis indicate the presence of amino acid residues surrounding and interacting with the Taxol binding site on β-tubulin, where molecular docking of the Avermectin compound has been conducted. The optimal docking configurations for each compound have been examined and are presented in separate tables. The results of the docking studies for each ligand within the classical tubulin binding sites of the 1TUB target are presented here.
Docking of ligands with 1TUB
The structural analysis of the 1TUB reveals that the α- and β-tubulin monomers, which constitute microtubules, are fundamentally similar. Each monomer is characterized by a core structure of two β-sheets flanked by α-helices and is segmented into three functional domains: The C-terminal domain: This domain predominantly forms the binding surface for motor proteins. The N-terminal domain: It contains the nucleotide-binding region. The intermediate domain: This domain includes the Taxol-binding site.
The primary Taxol binding site is located in the β-subunit, specifically near the top of helix H1 (between residues 15 and 25) and close to helix H5 and the H5-H6 loop (between residues 212 and 222). The principal interaction of the taxane ring with tubulin occurs at the residue L275, positioned at the beginning of the B8-H9 loop [16].
The first site, which was selected for further analysis (referred to as site B) is situated at the junction between the two monomers, with involvement from residues Arg2 and Tyr36 of β-tubulin and Thr73 and Asn76 of α-tubulin. The second site (referred to as site A) is defined by specific residues including Val23, Asn26, Tyr36, His229, Ala233, Phe244 and Phe272 of β-tubulin, coinciding with the Taxol binding site [17].
The five best-docked configurations were selected based on better docking scores of each of the Avermectin compounds and the compared ligand Taxol, presented separately in tables. The docking analysis revealed that Ivermectin B1a has the highest binding affinity with almost equal binding affinity and higher than Taxol for β-tubulin based on the docking energy and number of H-bonds followed by Selamectin and Doramectin, the lowest compound was Avermectin B1a with -8.2 Kcal/mol. All details of the atoms involved in bonding with ligands, bond lengths, docking energies and Ki values are given in tables.
In a hypothetical scenario, if the affinity of a ligand for its target protein is higher, its activity at the cellular or organism level would likely be more effective. Consequently, Ivermectin B1a, could potentially exhibit greater effectiveness compared to Taxol, -18.0 Kcal/mol, followed by Selamectin, Doramectin, Milbemycin oxime, Eprinomectin B1a and Moxidectin, respectively. These results are in good agreement with the in vitro experimental studies of Ivermectin with HeLa cell lines [13].
Moreover, the Milbemycin oxime, Eprinomectin B1a, Moxidectin and Avermectin B1a, have high binding affinity, with -8.8, -8.6, -8.6 and -8.2 Kcal/mol respectively. The Taxol binding affinity to β-tubulin was 17.4Kcal/mol. These findings align well with in vitro experimental studies involving Avermectin B1a and HCT-116 cell lines [11].
Similarly, in a hypothetical interaction between Avermectin compounds and β-tubulin, cellular functions might be impaired due to the inhibition of microtubule formation or alteration in microtubule stability. However, further assessment of the efficacy of these compounds is necessary.
Docking ofIvermectinB1a
For the binding of IvermectinB1a with the 1TUB, the highest observed binding affinity is -18.0 Kcal/mol. It has the highest binding energy in comparison to the binding of other Avermectin compounds considered for the study. The docking analysis revealed that Ivermectin B1a forms strong hydrogen bonds with key residues within the Taxol-binding site of β-tubulin, specifically residues such as Val23, His229, Ala233 and Phe270 (Figures 2a and 2b). These residues are important for stabilizing the α/β-tubulin dimer, making them critical in the process of microtubule assembly. The binding affinity of Ivermectin B1a is higher than that of Taxol (-17.4 Kcal/mol), indicating that it may act as a potent microtubule stabilizer. This suggests that Ivermectin could impede cancer cell proliferation by preventing tubulin from disassembling, similar to how Taxol operates, but potentially with greater efficacy, as detailed in Table 1.
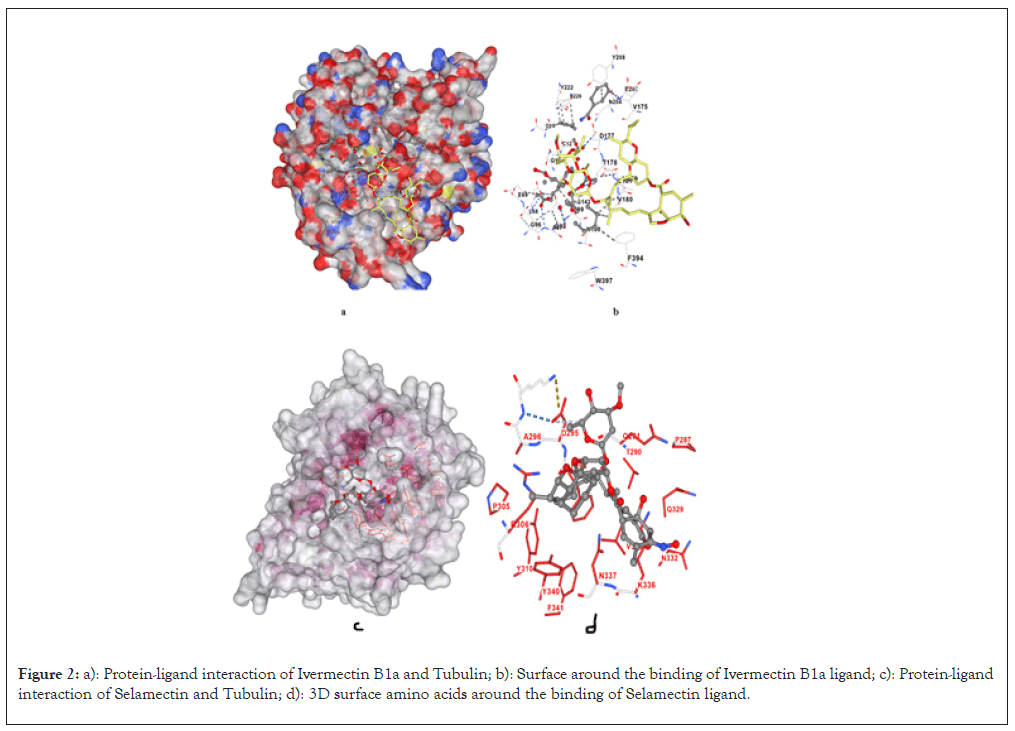
Figure 2: a): Protein-ligand interaction of Ivermectin B1a and Tubulin; b): Surface around the binding of Ivermectin B1a ligand; c): Protein-ligand interaction of Selamectin and Tubulin; d): 3D surface amino acids around the binding of Selamectin ligand.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume ( A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C1 | -18 | 3393 | 312, 439, 263 | 31, 31, 31 | Chain B: GLY10 GLN11 CYS12 GLN15 ILE16 ASP67 LEU68 GLU69 PRO70 GLN94 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 LYS103 GLU108 SER138 GLY141 GLY142 THR143 GLY144 VAL169 LYS174 VAL175 SER176 ASP177 THR178 VAL179 VAL180 GLU181 ASN204 GLU205 TYR208 TYR222 ASN226 GLN384 PHE394 TRP397 GLU401 |
| C2 | -16.6 | 743 | 324, 424, 280 | 31, 31, 31 | Chain B: LYS19 GLU22 VAL23 SER25 ASP26 GLU27 GLY29 ILE30 ASP31 PRO32 HIS37 PRO80 PHE81 LEU215 LEU217 THR221 ASP224 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 ARG318 PRO358 ARG359 GLY360 LEU361 |
| C4 | -13.9 | 191 | 329, 443, 292 | 31, 31, 31 | Chain B: ALA206 ASP209 ARG213 THR214 LEU273 TYR281 ARG282 ALA283 THR285 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 MET299 ALA302 ASP304 PRO305 ARG306 HIS307 TYR310 GLN329 ASN332 VAL333 ASN335 LYS336 ASN337 TYR340 PHE341 |
| C5 | -13.3 | 159 | 304, 457, 262 | 31, 31, 31 | Chain B: TYR183 LYS379 SER382 GLU383 GLN384 PHE385 THR386 ALA387 PHE389 ARG390 ARG391 LYS392 LEU395 THR399 GLY400 MET403 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 VAL419 |
| C3 | -12.9 | 207 | 295, 449, 268 | 31, 31, 31 | Chain B: TRP101 HIS105 TYR106 THR107 GLU108 ALA110 GLU111 VAL113 ASP114 LEU150 SER153 LYS154 ARG156 GLU157 PRO160 LEU187 HIS190 GLN191 GLU194 ASN195 GLY400 GLU401 GLY402 MET403 MET406 GLU407 PHE408 GLU410 ALA411 SER413 ASN414 |
Table 1: CB-Dock results for Ivermectin B1a-Tubulin interaction.
Docking of Selamectin
Selamectin binds to the Taxol-binding site with binding affinity of -9.1 Kcal/mol, forming hydrogen bonds with residues such as Asp26, Gly223, Thr221 and Leu228. These interactions occur at sites involved in the lateral contacts between α and β-tubulin subunits (Figures 2c and 2d). Selamectin’s ability to bind in the same region as Taxol, coupled with its high binding affinity, suggests that it could inhibit microtubule depolymerization, leading to cell cycle arrest and apoptosis in cancer cells, as outlined in Table 2.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C2 | -9.1 | 743 | 324, 424, 280 | 26, 26, 26 | Chain B: GLU22 VAL23 SER25 ASP26 PHE81 CYS211 LEU215 LYS216 LEU217 ASP224 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 GLN280 LEU284 PRO358 ARG359 GLY360 LEU361 LYS362 |
| C1 | -8.2 | 2626 | 312, 439, 266 | 26, 26, 26 | Chain B: GLY10 GLN11 CYS12 ASP67 LEU68 GLU69 GLY71 THR72 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY141 GLY142 THR143 GLY144 SER176 ASP177 THR178 VAL179 VAL180 GLU181 ASN204 TYR208 TYR222 PHE394 TRP397 |
| C4 | -7.9 | 191 | 329, 443, 292 | 26, 26, 26 | Chain B: ARG213 THR214 LYS216 LEU273 SER275 SER278 ARG282 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 ARG306 TYR310 GLU328 GLN329 ASN332 VAL333 LYS336 ASN337 TYR340 |
| C3 | -7.7 | 207 | 295, 449, 268 | 26, 26, 26 | Chain B: TRP101 HIS105 TYR106 THR107 ALA110 GLU111 VAL113 ASP114 LEU150 SER153 LYS154 ARG156 GLU157 HIS190 GLN191 GLU194 ASN195 ARG262 GLU401 GLY402 MET403 ASP404 MET406 GLU407 GLU410 ASN414 |
| C5 | -7.7 | 159 | 304, 457, 262 | 26, 26, 26 | Chain B: TYR183 LYS379 SER382 GLU383 PHE385 THR386 ALA387 PHE389 ARG390 ARG391 LYS392 LEU395 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 ASP417 |
Table 2: CB-Dock results for Selamectin-Tubulin interaction.
Docking of doramectin
For the binding of Doramectin with the 1TUB, the high-observed binding affinity is -8.9 Kcal/mol. It has the third-best binding energy following Ivermectin B1a. Doramectin binds to the Taxol-binding pocket through hydrogen bonding with residues such as Leu217, Thr221, His227 and Phe244. These interactions occur in regions critical for tubulin dynamics, particularly where Taxol is known to exert its stabilizing effects (Figures 3a and 3b). With a binding score almost equal to Milbemycin oxime, Doramectin demonstrates a similar potential to act as a microtubule-stabilizing agent. Given the comparable binding affinity, Doramectin could serve as an alternative to Taxol in targeting β-tubulin in cancer cells, which could offer therapeutic advantages depending on its pharmacokinetic profile, as detailed in Table 3.
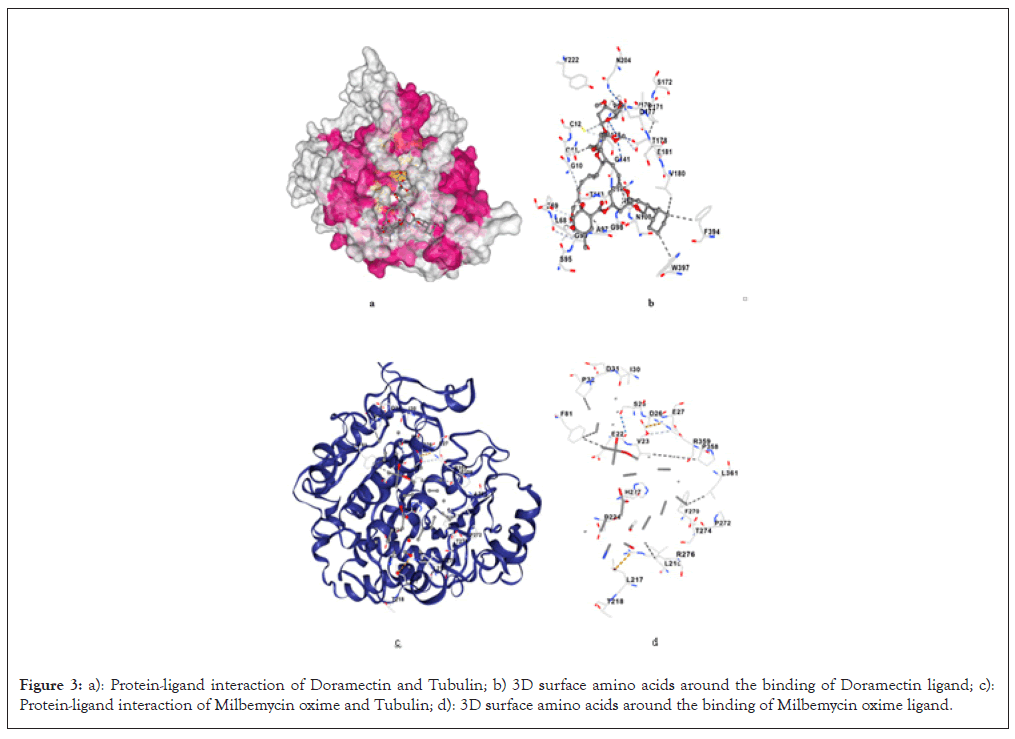
Figure 3: a): Protein-ligand interaction of Doramectin and Tubulin; b) 3D surface amino acids around the binding of Doramectin ligand; c): Protein-ligand interaction of Milbemycin oxime and Tubulin; d): 3D surface amino acids around the binding of Milbemycin oxime ligand.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C1 | -8.9 | 2626 | 312, 439, 266 | 30, 30, 30 | Chain B: GLY10 GLN11 CYS12 GLN15 ASP67 LEU68 GLU69 THR72 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY140 GLY141 GLY142 THR143 GLY144 VAL169 VAL170 PRO171 SER172 PRO173 LYS174 VAL175 SER176 ASP177 THR178 VAL179 VAL180 GLU181 PRO182 ASN204 GLU205 TYR208 TYR222 GLN384 MET388 PHE394 TRP397 TYR398 |
| C2 | -8.3 | 743 | 324, 424, 280 | 30, 30, 30 | Chain B: GLN15 ALA18 LYS19 GLU22 VAL23 SER25 ASP26 GLU27 ILE30 ASP31 PRO32 SER75 VAL76 PRO80 PHE81 CYS211 LEU215 LEU217 THR218 THR219 THR221 TYR222 GLY223 ASP224 LEU225 ASN226 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 GLN280 PRO358 ARG359 GLY360 LEU361 |
| C4 | -8.3 | 191 | 329, 443, 292 | 30, 30, 30 | Chain B: ARG213 THR214 LYS216 LEU273 SER275 SER278 TYR281 ARG282 ALA283 LEU284 THR285 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 ARG306 HIS307 GLN329 VAL333 LYS336 ASN337 TYR340 |
| C5 | -7.6 | 159 | 304, 457, 262 | 30, 30, 30 | Chain B: TYR183 HIS190 LYS379 SER382 GLU383 PHE385 THR386 PHE389 ARG390 ARG391 LYS392 ALA393 PHE394 LEU395 GLU405 MET406 PHE408 THR409 GLU410 GLU412 SER413 ASN414 ASN416 ASP417 VAL419 |
| C3 | -7 | 207 | 295, 449, 268 | 30, 30, 30 | Chain B: HIS105 TYR106 THR107 ALA110 GLU111 LEU112 VAL113 ASP114 LEU117 ARG121 LEU150 SER153 LYS154 ARG156 GLU157 GLU158 PRO160 HIS190 GLN191 GLU194 ASN195 ARG262 GLU401 GLY402 MET403 ASP404 MET406 GLU407 GLU410 SER413 ASN414 ASP417 |
Table 3: CB-Dock results for Doramectin-β-Tubulin interaction.
Docking of milbemycin oxime
The binding affinity of Milbemycin oxime was shown to be -8.8 Kcal/mol. Milbemycin oxime shows interactions with residues Val23, Phe81, His227 and Phe270, similar to other Avermectin family compounds. These residues are involved in the stabilization of the tubulin dimer. Milbemycin oxime's binding affinity, although slightly lower than Ivermectin, is still superior to some of the other compounds (Figures 3c and 3d). It indicates potential utility in disrupting microtubule functions, making it a candidate for further exploration in cancer treatment, as detailed in Table 4.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C2 | -8.8 | 743 | 324, 424, 280 | 31, 31, 31 | Chain B: LYS19 GLU22 VAL23 SER25 ASP26 GLU27 ILE30 ASP31 PRO32 THR33 GLY79 PRO80 PHE81 GLN83 LEU215 LEU217 THR219 PRO220 THR221 ASP224 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 ARG318 PRO358 ARG359 GLY360 LEU361 SER364 |
| C1 | -7.4 | 2626 | 312, 439, 266 | 31, 31, 31 | Chain B: GLY10 GLN11 CYS12 ASN14 GLN15 GLU69 GLY71 THR72 MET73 SER75 VAL76 SER95 GLY96 GLY98 ASN99 ASN100 LYS103 GLU108 SER138 GLY141 GLY142 THR143 GLY144 SER145 VAL169 VAL175 SER176 ASP177 THR178 VAL179 VAL180 GLU181 ASN204 GLU205 TYR208 PRO220 THR221 TYR222 PHE394 HIS396 TRP397 TYR398 |
| C4 | -6.9 | 191 | 329, 443, 292 | 31, 31, 31 | Chain B: THR214 SER275 TYR281 ARG282 ALA283 LEU284 THR285 PRO287 GLU288 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ARG306 HIS307 GLU325 GLU328 GLN329 ASN332 ASN335 LYS336 ASN337 TYR340 |
| C5 | -6.8 | 159 | 304, 457, 262 | 31, 31, 31 | Chain B: TYR183 GLU376 LYS379 ARG380 SER382 GLU383 PHE385 THR386 ALA387 PHE389 ARG390 ARG391 LYS392 ALA393 PHE394 LEU395 HIS396 TRP397 THR399 GLY400 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 ASP417 VAL419 SER420 |
| C3 | -6.6 | 207 | 295, 449, 268 | 31, 31, 31 | Chain B: HIS105 TYR106 THR107 GLU108 ALA110 GLU111 LEU112 VAL113 ASP114 LEU117 LEU150 SER153 LYS154 GLU157 GLU158 HIS190 GLN191 ASN195 THR399 GLY400 GLU401 GLY402 MET403 ASP404 GLU407 GLU410 |
Table 4: CB-Dock results for Milbemycin oxime β-Tubulin interaction.
Docking of moxidectin
For the binding of Moxidectin with the 1TUB, the observed binding affinity is -8.6 Kcal/mol. Moxidectin interacts with residues Glu22, Val23, Ser25 and Asp26 in β-tubulin. These residues are known to contribute to the dynamic instability of microtubules, which is essential for their rapid assembly and disassembly during cell division. Moxidectin’s strong binding affinity indicates its potential to stabilize tubulin, similar to Taxol, but possibly with different pharmacodynamics (Figures 4a and 4b). Its docking energy suggests it may have robust anticancer properties, but is lower than Ivermectin B1a and Doramectin, as detailed in Table 5.
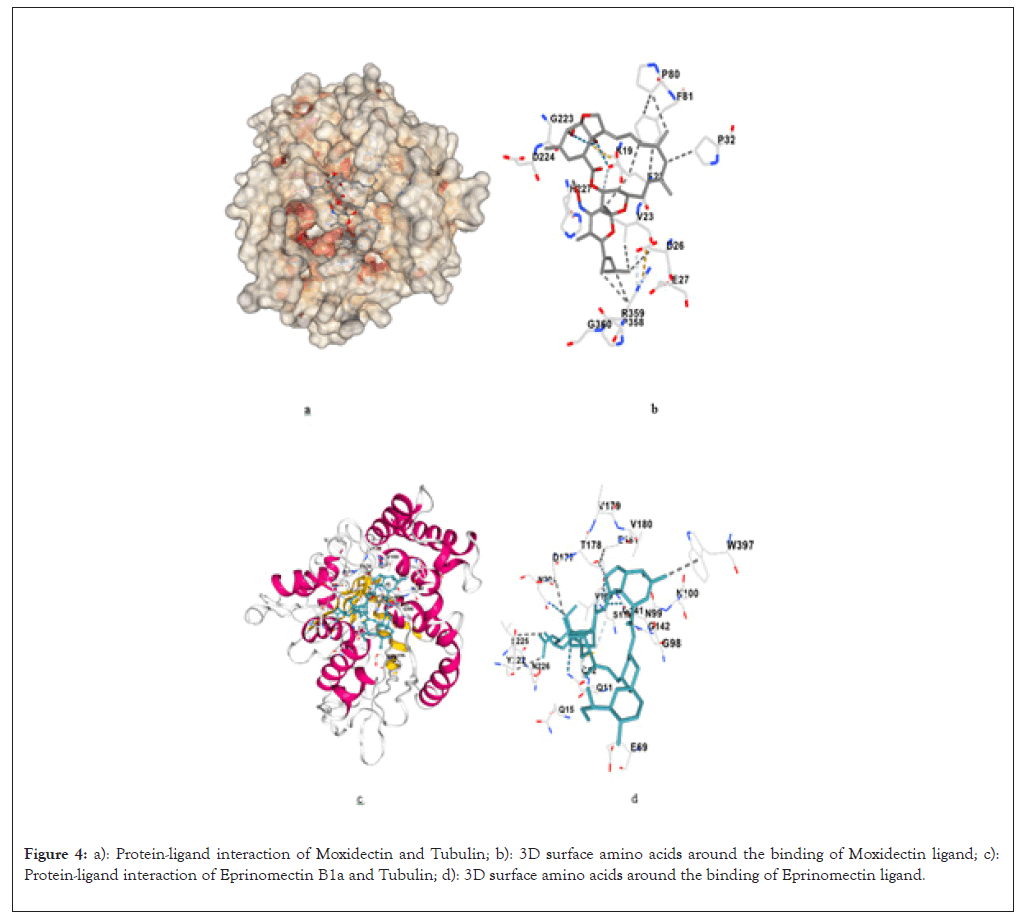
Figure 4: a): Protein-ligand interaction of Moxidectin and Tubulin; b): 3D surface amino acids around the binding of Moxidectin ligand; c): Protein-ligand interaction of Eprinomectin B1a and Tubulin; d): 3D surface amino acids around the binding of Eprinomectin ligand.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C2 | -8.6 | 743 | 324, 424, 280 | 25, 25, 25 | Chain B: GLN15 LYS19 GLU22 VAL23 SER25 ASP26 GLU27 PRO32 PRO80 PHE81 CYS211 LEU215 LYS216 LEU217 THR219 THR221 GLY223 ASP224 LEU225 HIS227 LEU228 ALA231 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 PRO358 ARG359 GLY360 LEU361 |
| C1 | -7.7 | 2626 | 312, 439, 266 | 25, 25, 25 | Chain B: GLY10 GLN11 CYS12 GLN15 ILE16 ASP67 LEU68 GLU69 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY140 GLY141 GLY142 THR143 GLY144 VAL169 VAL170 PRO171 SER172 VAL175 SER176 ASP177 THR178 VAL179 GLU181 ASP203 ASN204 GLU205 TYR208 TYR222 LEU225 ASN226 PHE394 |
| C4 | -6.9 | 191 | 329, 443, 292 | 25, 25, 25 | Chain B: ARG213 THR214 LEU273 ARG282 THR285 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 ARG306 TYR310 GLN329 ASN332 VAL333 ASN335 LYS336 ASN337 TYR340 PHE341 |
| C5 | -6.8 | 159 | 304, 457, 262 | 25, 25, 25 | Chain B: TYR183 PHE385 THR386 PHE389 ARG390 ARG391 LYS392 LEU395 HIS396 TRP397 THR399 GLY400 MET403 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 |
| C3 | -6.7 | 207 | 295, 449, 268 | 25, 25, 25 | Chain B: TRP101 HIS105 TYR106 THR107 GLU108 ALA110 GLU111 VAL113 ASP114 LEU150 SER153 LYS154 GLU157 HIS190 GLN191 GLU194 ASN195 GLU401 GLY402 MET403 ASP404 MET406 GLU407 GLU410 ASN414 |
Table 5: CB-Dock results for Moxidectin-Tubulin interactions.
Docking of eprinomectin
For the binding of Eprinomectin with the 1TUB, the observed binding affinity is -8.6 Kcal/mol. Eprinomectin interacts with residues Ser25, Leu215, His227 and Phe270 of β-tubulin. These residues are involved in stabilizing the α/β-tubulin interface, which is essential for maintaining the integrity of microtubules during cellular processes like mitosis. The binding energy of Eprinomectin is on par with Doramectin, which suggests that it could also stabilize microtubules effectively (Figures 4c and 4d). This compound’s similar binding profile to both Ivermectin B1a and Doramectin highlights its potential as an anticancer agent, as outlined in Table 6.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C1 | -8.6 | 2626 | 312, 439, 266 | 30, 30, 30 | Chain B: GLY10 GLN11 CYS12 GLN15 ILE16 ASP67 LEU68 GLU69 GLY71 THR72 SER75 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY141 GLY142 THR143 GLY144 VAL169 VAL175 ASP177 THR178 VAL179 VAL180 GLU181 ASN204 GLU205 TYR208 PRO220 THR221 TYR222 LEU225 ASN226 PHE394 TRP397 |
| C2 | -8.6 | 743 | 324, 424, 280 | 30, 30, 30 | Chain B: GLN15 ALA18 LYS19 GLU22 VAL23 SER25 ASP26 GLU27 ILE30 ASP31 PRO32 GLN43 VAL76 PRO80 PHE81 CYS211 LEU215 LYS216 LEU217 THR221 GLY223 ASP224 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 GLN280 ARG282 LEU284 ARG318 ARG320 ILE356 PRO357 PRO358 ARG359 GLY360 LEU361 LYS362 |
| C5 | -8.3 | 159 | 304, 457, 262 | 30, 30, 30 | Chain B: TYR183 LYS379 ARG380 SER382 GLU383 PHE385 THR386 ALA387 PHE389 ARG390 ARG391 LYS392 LEU395 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 VAL419 |
| C3 | -7.7 | 207 | 295, 449, 268 | 30, 30, 30 | Chain B: TRP101 HIS105 TYR106 THR107 ALA110 GLU111 VAL113 ASP114 LEU150 SER153 LYS154 ARG156 GLU157 HIS190 GLN191 GLU194 ASN195 ARG262 GLU401 GLY402 MET403 ASP404 MET406 GLU407 GLU410 ASN414 |
| C4 | -7 | 191 | 329, 443, 292 | 30, 30, 30 | Chain B: ARG213 THR214 LEU273 SER278 TYR281 ARG282 ALA283 THR285 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 ASP304 ARG306 HIS307 TYR310 VAL333 LYS336 ASN337 SER339 TYR340 |
Table 6: CB-Dock results for Eprinomectin B1a-Tubulin interaction.
Docking of avermectin B1a
The binding affinity of Avermectin B1a was shown to be -8.2 Kcal/mol and the interaction site of Avermectin B1a binds with residues like Glu22, Val23 and His227, interacting with regions critical for microtubule stability. It engages in similar interactions as other Avermectins in this family, although its binding energy is slightly lower (Figures 5a and 5b).
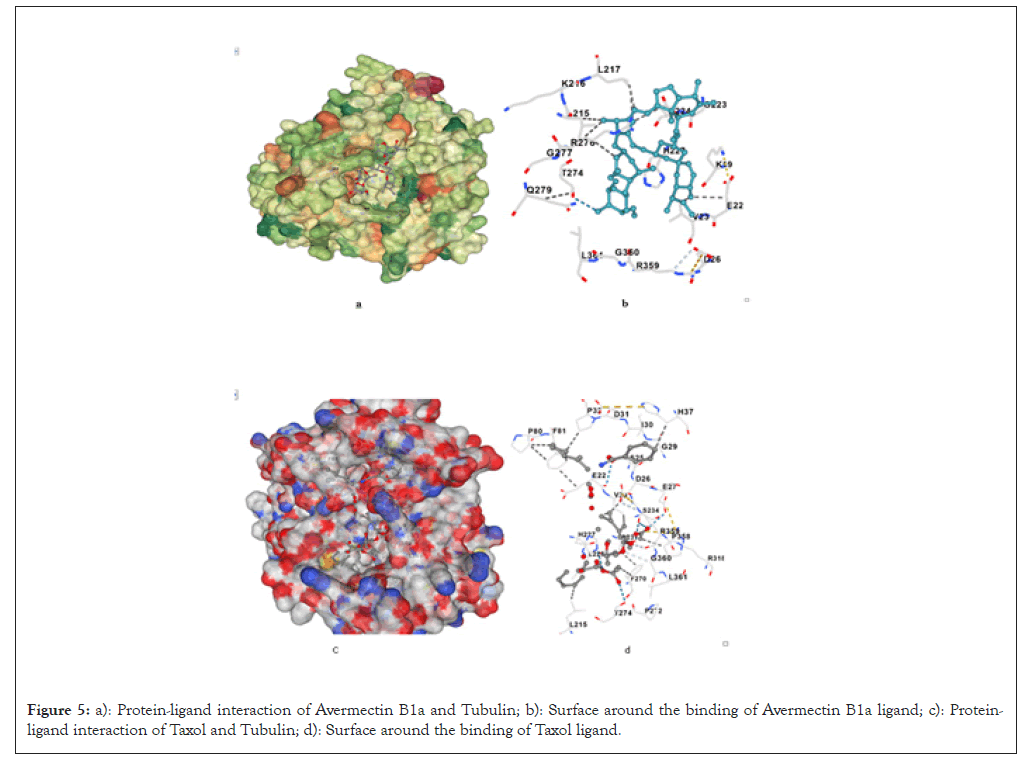
Figure 5: a): Protein-ligand interaction of Avermectin B1a and Tubulin; b): Surface around the binding of Avermectin B1a ligand; c): Protein-ligand interaction of Taxol and Tubulin; d): Surface around the binding of Taxol ligand.
Despite having a lower binding affinity than Ivermectin, Doramectin and Eprinomectin, Avermectin B1a still exhibits significant binding energy, indicating that it could disrupt tubulin dynamics and potentially act as an anti-mitotic agent in Table 7.
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C2 | -8.2 | 743 | 324, 424, 280 | 27, 27, 27 | Chain B: LYS19 GLU22 VAL23 SER25 ASP26 GLU27 PRO32 PRO80 PHE81 LEU215 LYS216 LEU217 THR221 GLY223 ASP224 HIS227 LEU228 ALA231 SER234 PHE270 PRO272 LEU273 THR274 SER275 ARG276 GLY277 GLN279 GLN280 PRO358 ARG359 GLY360 LEU361 SER364 |
| C5 | -8.1 | 159 | 304, 457, 262 | 27, 27, 27 | Chain B: TYR183 SER382 PHE385 THR386 PHE389 ARG390 ARG391 LYS392 LEU395 ASP404 GLU405 MET406 PHE408 THR409 GLU412 SER413 ASN416 |
| C1 | -8 | 2626 | 312, 439, 266 | 27, 27, 27 | Chain B: GLY10 GLN11 CYS12 GLY13 GLN15 ASP67 LEU68 GLU69 THR72 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY141 GLY142 THR143 GLY144 LYS174 VAL175 SER176 ASP177 THR178 VAL179 VAL180 GLU181 ASN204 GLU205 TYR208 TYR222 |
| C3 | -7.5 | 207 | 295, 449, 268 | 27, 27, 27 | Chain B: TRP101 HIS105 TYR106 ALA110 GLU111 VAL113 ASP114 LEU150 SER153 LYS154 ARG156 GLU157 HIS190 GLN191 GLU194 ASN195 MET403 ASP404 GLU407 GLU410 SER413 ASN414 |
| C4 | -7.2 | 191 | 329, 443, 292 | 27, 27, 27 | Chain B: ARG213 THR214 ARG282 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 ARG306 TYR310 GLN329 ASN332 VAL333 LYS336 ASN337 SER339 TYR340 |
Table 7: CB-Dock results for AvermectinB1a with β -Tubulin interaction.
Docking of taxol
Taxol binding with the 1TUB, the highest observed binding affinity is -17.4 Kcal/mol. In the optimal binding configuration with 1TUB, Taxol is involved in hydrogen bonding interactions of the O-H…O and N-H…O types with specific amino acid residues, as detailed in Table 8 (Figures 5c and 5d).
| Cur pocket ID | Vina score Kcal/mol | Cavity volume (A3) | Center (x,y,z) | Docking size (x,y,z) | Contact residues |
|---|---|---|---|---|---|
| C2 | -17.4 | 743 | 324, 424, 280 | 31, 31, 31 | Chain B: LYS19 GLU22 VAL23 SER25 ASP26 GLU27 HIS28 GLY29 ILE30 ASP31 PRO32 HIS37 GLN43 PRO80 PHE81 CYS211 LEU215 LEU217 THR221 GLY223 ASP224 HIS227 LEU228 ALA231 SER234 PHE242 PHE270 PRO272 THR274 SER275 ARG276 GLY277 GLN279 ARG318 ILE356 PRO357 PRO358 ARG359 GLY360 LEU361 |
| C1 | -15.8 | 2626 | 312, 439, 266 | 31, 31, 31 | Chain B: ALA9 GLY10 GLN11 CYS12 GLN15 ILE16 ASP67 LEU68 GLU69 GLY71 THR72 SER75 SER95 GLY96 ALA97 GLY98 ASN99 ASN100 SER138 GLY141 GLY142 THR143 GLY144 SER145 VAL169 VAL170 VAL175 ASP177 THR178 VAL179 VAL180 GLU181 ILE202 ASN204 GLU205 LEU207 TYR208 TYR222 LEU225 ASN226 VAL229 PHE394 TRP397 TYR398 |
| C4 | -13.8 | 191 | 329, 443,292 | 31, 31, 31 | Chain B: ARG213 THR214 LEU273 TYR281 ARG282 ALA283 THR285 PRO287 GLU288 THR290 GLN291 GLN292 PHE294 ASP295 ALA296 LYS297 ASN298 PRO305 ARG306 TYR310 GLN329 ASN332 VAL333 ASN335 LYS336 ASN337 TYR340 PHE341 |
| C5 | -12.7 | 159 | 304, 457, 262 | 31, 31, 31 | Chain B: TYR183 LYS379 ARG380 SER382 GLU383 PHE385 THR386 ALA387 PHE389 ARG390 ARG391 LYS392 LEU395 GLU405 MET406 PHE408 THR409 GLU410 GLU412 SER413 ASN416 ASP417 VAL419 SER420 |
| C3 | -12.2 | 207 | 295, 449, 268 | 31, 31, 31 | Chain B: TRP101 HIS105 TYR106 THR107 GLU108 ALA110 GLU111 VAL113 ASP114 LEU117 ARG121 LEU150 SER153 LYS154 ARG156 GLU157 GLU158 LEU187 HIS190 GLN191 GLU194 ASN195 ARG262 GLY400 GLU401 GLY402 MET403 ASP404 MET406 GLU407 PHE408 GLU410 ALA411 SER413 ASN414 |
Table 8: CB-Dock results for Taxol-Tubulin interaction.
Comparative insights
Ivermectin B1a stands out with the highest binding affinity (-18.0 Kcal/mol), suggesting it has the most potent interaction with β-tubulin. This supports its candidacy for further development as a tubulin-targeting agent.
Selamectin, Doramectin, Milbemycin oxime, Eprinomectin and Moxidectin share very similar binding profiles and docking energies (-9.1, -8.9, -8.8, -8.6, -8.6 Kcal/mol), suggesting they could all serve as effective alternatives to Taxol in targeting microtubules.
Avermectin B1a exhibits slightly lower binding affinities (-8.2 Kcal/mol) but still outperforms the binding energy, reinforcing the potential of Avermectin derivatives as microtubule-stabilizing agents. Previous studies support the anti-mitotic activity of Avermectin compounds through microtubule stabilization, such as the work by who observed that Ivermectin stabilizes mammalian microtubules similarly to Taxol in vitro, resulting in the inhibition of cancer cell proliferation in HeLa cells [13]. These findings align with the strong binding affinities observed here, with Ivermectin B1a showing the highest affinity (-18.0 Kcal/mol) among the tested Avermectin compounds, highlighting its potential as a potent microtubule-stabilizing agent.
Additionally, several Avermectin derivatives have been investigated for anticancer effects, with Ivermectin and Abamectin reported to inhibit tumor growth by disrupting microtubule dynamics [6]. The comparable affinities observed in our study suggest that other Avermectins, including Doramectin and Selamectin, might exhibit similar anti-proliferative effects due to their binding interactions with critical β-tubulin residues. Selamectin and Doramectin both achieved binding affinities (-9.1 and -8.9 Kcal/mol, respectively) close to Ivermectin, suggesting they could also serve as potential alternatives to Taxol, particularly in cases where Taxol-resistant cancers require novel therapeutic strategies.
The molecular interactions observed here with key β-tubulin residues, such as Val23, His227 and Phe270 are consistent with prior research on Taxol binding. For instance, characterized the Taxol binding pocket on β-tubulin, identifying these residues as critical to the stabilization of microtubules, which is essential for anti-mitotic activity [16]. Our results extend these findings by showing that Avermectin compounds interact with these same residues, reinforcing the concept that Avermectins could mimic Taxol’s action on microtubule stabilization but with potentially varied pharmacokinetics and resistance profiles.
In recent years, other studies have shown that Avermectin derivatives like Ivermectin also modulate additional cellular pathways, such as the Akt/mTOR pathway [10]. Such modulation can enhance apoptosis and hinder cell proliferation in cancer cells, indicating that Avermectins might exhibit multi-target activity beneficial for overcoming cancer cell resistance mechanisms. While this study focuses on molecular docking and affinity with β-tubulin, future work should explore how these interactions translate into in vitro efficacy, considering that Avermectins might provide both direct and indirect inhibition of cancer cell growth through multi-pathway modulation.
In conclusion, using molecular docking, we demonstrated the interactions of the natural derivative compounds of the Avermectin family with β-tubulin. Our results show that Avermectin compounds, including Ivermectin B1a, Selamectin and Doramectin, show a positive ability to bind to β-tubulin with binding affinities equal to or surpassing Taxol.
Overall, these findings position Avermectin compounds as strong candidates for development as tubulin-targeting agents, with binding affinities that could support their use as alternatives to Taxol in cancer treatment. Comparative binding data suggest that the microtubule-stabilizing potential of Avermectins, particularly Ivermectin B1a, Selamectin and Doramectin, warrants further investigation to elucidate there in vivo efficacy and possible advantages over taxane-based drugs, such as reduced toxicity or better pharmacodynamics in resistant cancer types.
All authors hold equal contribution.
All authors declare no conflict of interest.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Hoti Q, Ghali UM, Adem E, Hoti K, Gavazaj F (2024). Investigating the Interaction of Avermectin Family Compounds against Tubulin Protein: A Molecular Docking Study. J Theor Comput Sci. 10:227.
Received: 07-Nov-2024, Manuscript No. JTCO-24-35028; Editor assigned: 11-Nov-2024, Pre QC No. JTCO-24-35028 (PQ); Reviewed: 25-Nov-2024, QC No. JTCO-24-35028; Revised: 02-Dec-2024, Manuscript No. JTCO-24-35028 (R); Accepted: 09-Dec-2024 Published: 09-Dec-2024 , DOI: 10.35248/2376-130X.24.10.227
Copyright: © 2024 Hoti Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.