Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2022)Volume 12, Issue 5
Characterizing the fungal microbiota of free-living animals such as reptiles is necessary for a better understanding of fungal-host interactions as well as for establishing patterns of infections in these animals, thus creating an information network that helps understand the incidence and the microorganisms involved in this population for better maintenance and conservation of these species. Nonetheless, reports with this approach are limited, especially those focused on filamentous fungi. Therefore, this study aimed to isolate and identify filamentous fungi present in the oral cavity microbiota and cloaca of free-living reptiles of the Caatinga biome. Seven lizards, seven snakes, and two amphisbaenians were collected for mycological sampling, totaling 16 species of reptiles. The samples were collected from the oral and cloacal cavity of the animals, with sterile swabs, and sown in Sabouraud agar supplemented with chloramphenicol (0.05 g/L) and maintained for up to 14 days for the fungi isolation. Thus, 77 strains of filamentous fungi present in the microbiota of reptiles such as snakes, lizards, and amphisbaenian were isolated and identified. Among the most frequent fungi are the genera Aspergillus, Penicillium, and Microsporum, respectively. In short, knowledge of fungal diversity present in free-living reptiles of the Caatinga biome is of fundamental importance, since different opportunistic filamentous fungi have been isolated in this study, especially the genus Aspergillus, emphasizing the importance of monitoring these animals, as they can act as carriers and disseminators of potentially pathogenic agents.
Aspergillus sp.; Penicillium sp.; Fungal microbiota; Wild animals; Fungal diversity
The reptiles have suffered population declines caused by different emerging fungal pathogens over the last years [1]. In this context, although studies are limited, some diseases have already been reported and described, such as the gastrointestinal and pulmonary candidiasis in turtles [2] and ofidiomycosis, also known as “snake fungal disease”, caused by the filamentous fungus Chrysosporium ophiodiicola, first identified in 2009 [3].
To establish the infection and cause the disease, fungi express different virulence factors, which contribute to a greater pathogenicity. Castelo-Branco et al., highlighted the pathogenic potential of yeasts such as Candida famata isolated from the microbiota of reptiles and amphibians [4].
Studies describing the microbiota of animals are fundamental to understanding the microbiota-host interaction. In this regard, research characterizing the microbiota of reptiles has gained numbers in the literature [1,5-7]. Since bacteria and fungi associated or not with the animal microbiota may become pathogens under specific circumstances, these studies are of paramount importance for a better understanding of the biology and health of these animals, whereas intra and inter-species communication can play a major role in disease transmission [7]. Nevertheless, studies on the description and characterization of the filamentous fungal microbiota in wild reptile species are scarce [4-6]. Thus, this study aimed to isolate and identify filamentous fungi present in the oral cavity microbiota and cloaca of free-living reptiles.
Sampling
The animal sampling was performed in forests, where Caatinga biome could be found, in the municipality of Acaraú, CE, Brazil (2°52'28.6"S 40°06'26.1"W), through the active search method [8]. The total sampling area of the animals was of approximately 500,000 m2. The animals were handled according to law nº 11,794 of October 8, 2008, following all the established rules for handling and experimentation with live animals of the phylum Chordata. The collection of animals was authorized by the Institute of Biodiversity Chico Mendes (ICMBio) under the protocol number SISBIO-51820-1.
Samples were collected from 16 animals, five species of lizard (Ameiva ameiva, Ameivula ocellifera, Iguana iguana, Polychrus acutirostis and Tropidurus hispidus), seven species of snakes (Lygophis dilepis, Micrurus ibiboboca, Oxyrhopus trigeminus, Philodryas olfersii, Psomorphis jeobestis, Tantilla melanocephala e Xenodon merremii) and two aphisbaenians (Amphisbaena sp. e Amphisbaena vermicularis).
Sterile swabs were used to collect the microbiological material, which were inserted and rotated in the oral cavity and in the cloaca of the reptiles. The samples were transported in 2 mL of sterile saline solution (0.9%), refrigerated at 4°C, to the Laboratory of Environmental Biology and Microbiology (LABIAM), Federal Institute of Education, Science and Technology of Ceará-Campus Acaraú [9].
Sample processing and identification
The samples collected were inoculated in a 250 mm Petri dish containing Sabouraud Agar with 0.05 g/L chloramphenicol (Himedia Laboratories, Mumbai, India). Inoculation was performed with Swab used for sampling in direct contact with the culture medium through plaque depletion technique. A plate was used for each sample. After inoculation, the plates were kept at a temperature of 25-27°C for up to 14 days with daily observations [10].
After the fungal crops growth, they were described and isolated in Dextrose Potato Agar (Difco Laboratories, USA) and kept in the LABIAM mycotic library. A microcultivation was made in lamina using Dextrose Potato Agar (Difco Laboratories, USA) for observation of micromorphology and the ensuing identification of filamentous fungi, according to the De Hoog methodology (2002). The fungi were identified at the genus level and at the species level, whenever possible [11].
Statistical analysis
Non-parametric analysis was applied to compare the data. Thus, the Kruskal-Wallis’s test, followed by Dunn’s post hoc test, was applied for the comparison between the genera of the isolated fungi, as well as between the isolates from lizards, snakes, and amphisbaenians. Pearson’s correlation was applied to evaluate the correlation of isolates between animals. Frequency data were calculated so that absolute frequency (fa) corresponded to the total number of fungal isolates (fa=n), and relative frequency (fr) to the number of isolates of each genus (ng), and relative frequency (fr) to the number of isolates of each genus (ng) divided by fa (fr=ng/fa × 100). The maximum significance level adopted for affirmative conclusions was 5%.
The microorganisms were identified by morphological methods, as follows 77 filamentous fungi, belonging to the following genera: Alternaria, Aspergillus, Cladosporium, Cladophialophora, Fusarium, Hortaea, Microsporum, Mucor, Paelomyces, Penicillium, Rhizomucor, Rhizopus and Tricophyton (Table 1).
| Animal species | Anatomical site | Fungal species |
|---|---|---|
| Lizards | ||
| Ameiva ameiva | Oral cavity | Aspergillus sp. (1/77) * |
| Cladophialophora sp. (2/77) | ||
| Fusarium sp. (2/77) | ||
| Ameivula ocellifera | Oral cavity | Aspergillus sp. (2/77) |
| Hortaea werneckii (1/77) | ||
| Mucor sp. (2/77) | ||
| Rhizomucor sp. (1/77) | ||
| Rhizomucor variabilis (1/77) | ||
| Cloaca | Alternaria sp (2/77) | |
| Iguana iguana | Oral cavity | Aspergillus flavus (1/77) |
| Cladosporium sp. (2/77) | ||
| Penicillium sp. (2/77) | ||
| Cloaca | Aspergillus sp. (2/77) | |
| Aspergillus niger (3/77) | ||
| Cladosporium sp. (1/77) | ||
| Rhizomucor sp. (1/77) | ||
| Polychrus acutirostis | Oral cavity | Aspergillus sp. (1/77) |
| Aspergillus niger (1/77) | ||
| Cloaca | Aspergillus sp. (1/77) | |
| Aspergillus niger (1/77) | ||
| Microsporum sp. (1/77) | ||
| Penicillium sp. (1/77) | ||
| Tropidurus hispidus | Oral cavity | Aspergillus sp. (3/77) |
| Aspergillus niger (2/77) | ||
| Snakes | ||
| Lygophis dilepis | Oral cavity | Aspergillus sp.(1/77) |
| Mucor sp. (1/77) | ||
| Penicillium sp.(1/77) | ||
| Rhizopus sp.(1/77) | ||
| Cloaca | Aspergillus sp.(1/77) | |
| Micrurus ibiboboca | Oral cavity | Fusarium sp.(2/77) |
| Rhizomucor sp. (1/77) | ||
| Cloaca | Hortaea werneckii (2/77) | |
| Oxyrhopus trigeminus | Oral cavity | Penicillium sp. (3/77) |
| Cloaca | Aspergillus flavus (1/77) | |
| Aspergillus niger (2/77) | ||
| Microsporum sp. (1/77) | ||
| Microsporum gypsium (1/77) | ||
| Penicillium expansum (1/77) | ||
| Philodryas olfersii | Oral cavity | Alternaria sp. (2/77) |
| Aspergillus sp. (2/77) | ||
| Microsporum sp. (1/77) | ||
| Psomorphis jeobestis | Cloaca | Microsporum sp. (2/77) |
| Fusarium sp. (1/77) | ||
| Tantilla melanocephala | Oral cavity | Cladosporium sp. (1/77) |
| Xenodon merremii | Cloaca | Aspergillus sp. (2/77) |
| Paelomyces sp. (1/77) | ||
| Amphisbaenian | ||
| Amphisbaena sp. | Oral cavity | Aspergillus sp. (1/77) |
| Aspergisllus clavatus (1/77) | ||
| Cloaca | Cladophialophora sp. (1/77) | |
| Mucor sp. (2/77) | ||
| Penicillium sp. (1/77) | ||
| Amphisbaena vermicularis | Oral cavity | Trichophyton sp. (2/77) |
| Trichophyton interfigitalles (1/77) | ||
Table 1: Relationship of fungi isolated from animal species and their respective anatomical.
The highest fungal richness was associated with Iguana iguanaspecies, which was the most representative in number of fungal isolates among lizards, with 15.78% (12/77) of the isolates. Followed by the serpent Oxyrhopus trigeminus, with 11.69% (9/77), and finally amphisbaenian, Amphisbaena sp. with 7.79% (6/77) of the fungal isolates (Table 1).
Amongst the reptile taxa, the fungal genus Aspergillus was the most frequent among the species of lizards (48.64%) and snakes (29.03%), while among amphisbaenians the genus Trichophyton had a frequency of 33.33%. Considering the general distribution of fungi among all reptile species, Aspergillus was the most representative with a relative frequency of 37.66%, followed by Penicillium (11.68%) and Microsporum (7.79%) (p<0.0001) when compared to other genera (Figures 1 and 2).
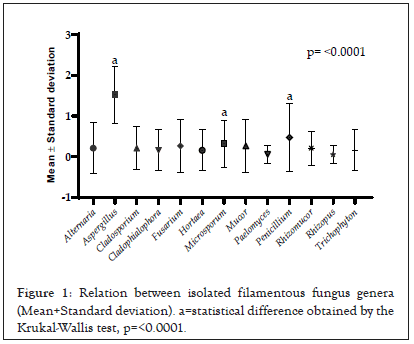
Figure 1: Relation between isolated filamentous fungus genera (Mean+Standard deviation). a=statistical difference obtained by the Krukal-Wallis test, p=<0.0001.
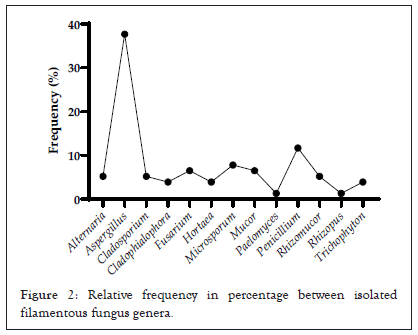
Figure 2: Relative frequency in percentage between isolated filamentous fungus genera.
Amongst the reptiles studied, lizards were the most representative in fungal isolates, with 37 isolates with p< 0.0001. Pearson correlation values of the animals were low, with r=-0.26 between lizards and snakes and r=0.11 between lizards and amphisbaenas.
Reports on filamentous fungi composing the microbiota of Caatinga reptiles are limited. The present study identified that reptiles may display present great diversity of filamentous fungi within their microbiota. In addition, most of the isolates found in this survey are known for being agents with pathogenic potential. According to Nardoni et al., fungi such as Aspergillus sp., Penicillium sp. and Fusarium sp. can be found in the cloaca of reptiles, which supports the findings of the present study [12].
In other studies, the genus Penicillium [6,12] was predominant amongst the remaining isolates. In the present study, the genus Aspergillus dominated the number of isolates. Despite some differences in frequency, the variety of isolates reported in this study is in agreement with other relevant findings regarding the fungal microbiota of reptiles [13]. The difference between the prevalence of isolates can probably be attributed to sampling methods, study design, variety of animal species, number of animals sampled, geographical location, among other factors.
The data from this research show that most reptile species share the same fungal isolates. The logic behind this sharing is uncertain, but may be related to common exogenous sources of contact with these fungi. Thus, the distribution of the same isolates in different hosts, as well as the presence of several fungal isolates in a single host, suggest that the reptiles might be acting as facultative transport animals of filamentous fungi.
Some studies report the pathogenic potential of fungi isolated from animals, such as reptiles [6,4,14]. Amongst the prevalent genera in this study, Aspergillus, followed by Penicillium and Microsporum, are correlated to diseases in animals and humans [15-18]. Aside from these, other isolated fungi with lower incidence such as Cladosporuim sp., Hortaea werneckii, Mucor sp. and Rhizomucor sp. have opportunistic pathogenic potential, and may cause numerous opportunistic infections in both animals and humans [19].
In conclusion, the knowledge of fungal diversity present in free-living reptiles of the Caatinga biome is fundamental. Different opportunistic filamentous fungi have been isolated in this study, especially the genus Aspergillus, emphasizing the importance of monitoring these animals, as they can serve as carriers and disseminators of potentially pathogenic agents. Data involving the isolation of filamentous fungi associated with free-living reptiles are scarce. The diversity associated with these wild animals, as well as the pathogenic potential of the genera isolated, demonstrated in this study, reveal the need and urge for research of this kind. In addition, such surveys help clarify questions about the microbiota of these animals, given the transient fungal microbiota.
Citation: Graça-Filho RV, Rocha MGD, Cunha Fonseca XMQ, Teixeira CF, Rocha Brandão AL, Neto Paiva MA (2022) Investigation of Filamentous Fungi in Wild Caatinga Biome Reptiles: Microbiota Research. Fungal Genom Biol. 12:198.
Received: 17-Aug-2022, Manuscript No. FGB-22-18880; Editor assigned: 22-Aug-2022, Pre QC No. FGB-22-18880 (PQ); Reviewed: 05-Sep-2022, QC No. FGB-22-18880; Revised: 12-Sep-2022, Manuscript No. FGB-22-18880 (R); Published: 19-Sep-2022 , DOI: 10.35841/2165-8056.22.12.198
Copyright: © 2022 Graça-Filho RV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.