Journal of Clinical and Experimental Ophthalmology
Open Access
ISSN: 2155-9570
ISSN: 2155-9570
Research Article - (2020)Volume 11, Issue 2
Objective: Glaucoma is characterized by a progressive degeneration of retinal ganglion cells (RGC) and their axons
leading to painless damage of the visual field and finally blindness. The exact pathophysiology of RGC loss remains
unknown.
Alterations in the microbiome may be linked to neurodegenerative conditions such as Alzheimer’s and Parkinson’s
diseases, possibly due to associated chronic low-grade inflammation. A recent study linked alterations in the oral
microbiome and glaucoma.
Methods: We investigated the microbiome of salivary and fecal samples in patients with normal tension glaucoma
(10), ocular hypertension (11) and controls (11) using a case-control design with 16S rDNA sequencing.
Results: For controls, but not the patient groups, salivary and fecal microbiome diversity was correlated in a given
patient, suggesting an uncoupled saliva and fecal microbiome in the diseased groups. Such findings suggest that
normal tension glaucoma (NTG) and ocular hypertension (OHT) might have similar characteristics. However, ocular
hypertension patients seem to be resistant to neurodegenerative disease progression indicating that the uncoupled
microbiome might affect characteristics linking ocular hypertension and normal tension glaucoma together.
Moreover, we found the salivary microbiome to contain more differential taxa-level abundances of microorganisms
suggesting the salivary microbiome might be advantageous to use in future studies investigating novel biomarkers in
ophthalmic neurodegenerative diseases.
Conclusion: The finding of an uncoupled microbiome might indicate comparable characteristics among glaucoma
patients and ocular hypertension patients.
Glaucoma; Microbiome; Ocular hypertension
Androgenic alopecia (AGA) (also known as pattern alopecia) is the most common form a chronic non-scarring hair loss, affecting both men and women [1]. AGA is characterized by progressive hair loss, especially of scalp hair, and has distinctive patterns of loss in women versus men, but in both genders the central scalp is most severely affected [2].
Follicular miniaturization and local high dihydrotestosterone (DHT) activity are considered the main pathological mechanisms involved in AGA [3]. Microinflammation is also a relevant pathogenetic mechanism involved in this clinical condition [4].
Approved therapeutic options are limited to topical minoxidil [5] for both men and women and oral finasteride [6] for men. However, both drugs possess side effects and are effective in less than 50% of treated patients [7].
Autologous Platelet-Rich Plasma (PRP) dermal injection is an additional therapeutic option for the medical treatment of AGA [8]. However, clinical efficacy of this approach in some cases could be disappointing [9]. Positive effects of PRP in AGA seem correlated, at least in part, with a growth factors mimicking action [10].
PRP is rich in growth factors like PDGF (platelet derived growth factor), TGF (transforming growth Factor Beta) and VEGF (Vascular endothelial growth Factor) [11]. A hair scalp lotion containing high-purified, growth factors-like polypeptides (octapeptide 2, acetyl decapeptide 3, oligopeptide 20 and copper tripeptide) with the addition of glycine and taurine (GFM-L) is available.
Copper tripeptide has shown to have antioxidant [12], anti inflammatory and blood vessel growth promoting action [13]. Copper tripeptide can increase the activity of FGF (Fibroblast Growth Factor) and VEGF [14]. Interestingly, copper tripeptide decreases the secretion of TGF-β [15] from dermal fibroblasts. TGF-β is involved in inducing catagen phase [16].
Copper tripeptide can also interfere with the activity of 5-α- reductase, therefore reducing the production of DHT [17]. Finally, this peptide is also able to stimulate the production of decorin [18].
Decorin, at scalp level, improves the anagen phase [19] and consequently hair growth [20]. The acetyl decapeptide is a synthetic peptide that mimics the action of two growth factors: the keratinocyte growth factor (KGF) and the epidermal growth factor (EGF), which acts on the follicle, promoting the anagen growth phase, through an antiapoptotic effect [21], maintaining the bulge stem cells active [22].
Another component of the growth factor like mixture, octapeptide 2, is able to promote hair growth [23], to reduce apoptosis and to increase keratinocyte proliferation [24].
In this formulation all the 4 oligopeptides are vehiculated in nanosomes of 250 nm in diameter. Published data state that nanosomes with a diameter<500 nm can transport molecules deep in the hair bulb, after topical application [25].
Taurine is a relevant amino acid with interesting hair growth promoting mechanisms [26]. This lotion has shown to be effective as stand-alone approach in the treatment of excessive hair loss in men and women [27].
The peculiar composition of this lotion (mixture of growth factor like peptides and taurine) suggests a potential synergistic effect on hair growth with other therapeutic strategies like PRP treatment. So far, no data regarding the potential synergistic effect of autologous PRP treatment combined with GFM-L are available.
We therefore conducted a prospective, randomized, assessorblinded trial to compare the efficacy and tolerability of PRP alone vs. PRP and GFM-L treatment in men and women with AGA.
Participants in the study
Ten patients with NTG and 11 patients with OHT were compared to 11 age- and gender-matched control subjects who had undergone an eye exam within 3 month, including visual acuity, slit lamp examination, IOP, funduscopy, visual field testing using Humphrey Field Analyzer (30:2 SITA Fast). The included patients with NTG had characteristic glaucomatous visual field losses and no untreated pressures measured above 16 mmHg. Patients with OHT had untreated IOP above 24 mmHg and no signs of glaucomatous damage.
All subjects enrolled in the study were caucasian females between 50 and 96 years of age. Subject were all non-smokers and with no alcohol abuse. They had no previous history of ocular trauma, no disease affecting the retina or the optic nerve, other than glaucomatous characteristics in the patients with NTG patients and increased IOP in patients with OHT. No subjects had significant systemic conditions. Moreover, subjects had no illness related to the gastrointestinal tract and had not used antibiotics three months prior to the collection of faeces and saliva.
The use of probiotics amongst the subjects was taken into consideration, but a more extensive registration of diet was not conducted. The characteristics of the participants enrolled are presented in table 1.
| Clinical data for group CTRL, NTG and OHT | |||
|---|---|---|---|
| Variable | CTRL (n=11) | NTG (n=10) | OHT (n=11) |
| Mean age | 67,7 | 70,3 | 71,5 |
| Ethnicity (white) | 1 | 1 | 1 |
| Females | 1 | 1 | 1 |
| BMI | 24,9 | 22,6 | 26,5 |
| Smoking | 0 | 0 | 0 |
| Alcohol* | 90,9% | 1 | 1 |
| Pro- or prebiotic intake | 18,2% | 11,1% | 9,1% |
Table 1: Characteristics of the participants enrolled in the study. *Alcohol within official limits.
Patient involvement
This research was carried out without involving patients in study design, writing of the manuscript or interpretation of the results.
Ophthalmic status
Every participant included in the study underwent ophthalmic examination a maximum of three months prior to the analysis of the saliva and faeces. Criteria for the NTG group: an untreated IOP of less than 16 mmHg, an open angle examined by gonioscopy, glaucomatous cupping of the optic nerve head, and characteristic visual field losses examined by Humphrey Field Analyzer on at least one eye. Criteria for the OHT group: Untreated IOP measurements above 24 mmHg, no signs of glaucoma including normal optic nerve heads, normal visual fields, as well as open angles examined by gonioscopy.
Samples
The samples were collected at home, frozen within five minutes of sampling to avoid bacterial overgrowth or degradation, transported on dry ice, and then kept at -20°C until analysis.
Written informed consent was obtained before the collection of samples. The study was performed in compliance with the Declaration of Helsinki and approved by the Danish National Ethical Committee (Ethical approval SJ-489).
DNA extraction
DNA was extracted from the fecal samples using the Nucleospin DNA extraction kit and from the saliva samples using proteinase K digestion [23].
PCR
PCR was done with the forward primer 515fB and the reverse primer 806rB with Illumina adaptors attached.
Illumina adapter and 515fB:
5'-
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGY
CAGCMGCCGC-GGTAA-3'
Illumina adapter and 806rB:
5'-
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGAC
TACNVGGGTWT-CTAAT-3'
These are universal bacterial 16S rDNA primers, which target the V4 region. The following PCR program was used: 98°C for 30 sec, 25x (98°C for 10 sec, 50°C for 20 sec, 72°C for 20 sec), 72°C for 5 min. Amplification was verified by running the products on an agarose gel. Barcodes were added in a subsequent PCR using the Nextera Index Kit V2 (Illumina) with the following PCR program: 98°C for 30 sec, 8x (98°C for 10 sec, 55°C for 20 sec, 72°C for 20 sec), 72°C for 5 min. Attachment of barcodes was verified by running the products on an agarose gel. Both initial and barcoding PCRs included negative controls.
Normalization and sequencing
Products from the nested PCR were pooled and cleaned using magnetic DNA-binding beads and the DNA concentration of pooled libraries was measured fluorometrically using a fluorescent DNA-binding dye. Sequencing was done on an Illumina MiSeq desktop sequencer (Illumina) using 2x250 bp chemistry.
Bioinformatical analysis
The 64-bit versions of USEARCH [24] and mother [25] were used in combination with several in-house programs for bioinformatical analysis of the sequencing data. Following tag identification and trimming, all sequences from all samples were pooled. A saliva sample from the NTG group failed to sequence, resulting in 9 patient samples from the NTG group where both saliva and faeces were sequenced.
Paired-end reads were merged requiring at least 30 bp overlap and a merged read length between 250 and 500 bp in length. Sequences with ambiguous bases, without perfect match to the primers or homopolymer length greater than 8, were discarded and primer sequences trimmed. Reads were quality filtered, discarding reads with more than 1 expected error and sequences strictly dereplicated, discarding clusters smaller than 5.
Sequences were clustered at 97% sequence similarity, using the most abundant strictly dereplicated reads as centroids and discarding suspected chimeras based on internal comparison. Additional suspected chimeric operational taxonomical units (OTUs) were discarded based on comparison with the Ribosomal Database Project classifier training set v9 [26] using UCHIME [27]. Taxonomic assignment of OTUs was done using the method by Wang et al. [28] with mothur's PDS version of the RDP training database v14. Following this, samples were rarified to the lowest sequence number found in a sample ≥1000 (after in silico removal of contaminating OTUs).
Statistical analysis
Statistical analyses were done in R.
Differences in the microbiomic composition were analyzed using the Kruskal-Wallis test as well as the Mann-Whitney U-test and corrected by multiple hypothesis testing using the Benjamini- Hochberg procedure by applying it for each taxonomical level for each sample type.
The diversity of the microbiome was assessed using the number of OTUs and the Shannon index, which in addition to the number of species also takes their relative abundance into account. To compare taxonomic composition between samples, UniFrac distances were used and compared by the Adonis nonparametric statistical method.
The diversity, measured by both number of OTUs and Shannon index, of a saliva sample was correlated with the diversity of the fecal sample from the same patient. Spearman correlations were calculated by subsetting to each group.
We explored whether two fecal samples from the same patients would be highly similar, if the two saliva samples were found to have highly similar microbiome composition, measured by their UniFrac distance. This was calculated using the Mantel test and done for all samples and by subsetting to each group.
Additionally, it was tested if the microbiome of a saliva sample were more similar to the fecal sample of the same patient than the fecal samples of other patients. This was calculated by the Mann-Whitney U test.
Microbiomic status of faeces and saliva in patient with NTG, patients with OHT, and healthy age- and gendermatched controls
To investigate the association between the gut microbiome and the glaucomatous loss of retinal ganglion cells, we collected salivary and fecal samples from 10 NTGs, 11 OHTs, and 11 ageand gender-matched controls.
The saliva samples were relatively diverse at class level in all three groups, while the fecal samples were completely dominated by the two classes of Clostridia and Bacteroidia (Figure 1).
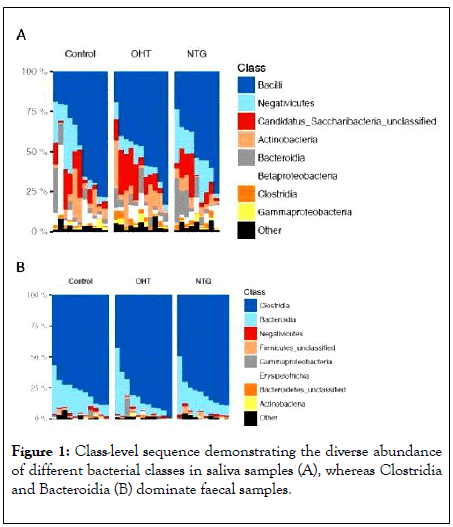
Figure 1: Class-level sequence demonstrating the diverse abundance of different bacterial classes in saliva samples (A), whereas Clostridia and Bacteroidia (B) dominate faecal samples.
Microbiomic status comparison between NTG patients and OHT patients
The microbiomic status between NTG patients and OHT patients showed some differential bacterial abundance between the two groups refered to as taxa (Figure 2). However, none of the differences were significant after correcting for multiple hypotheses testing by the Benjamini-Hochberg procedure to avoid the risk of false positive results. Before multiple hypotheses testing, the fecal abundances of Rikenellaceae and Alistipes and the salivary abundances of Negativicutes, Selenomonadales, Veillonella and Veillonellacea were found to be significant higher in the NTG group compared to the OHT group. All other comparisons of the taxa-level abundances between the groups were non-significant.
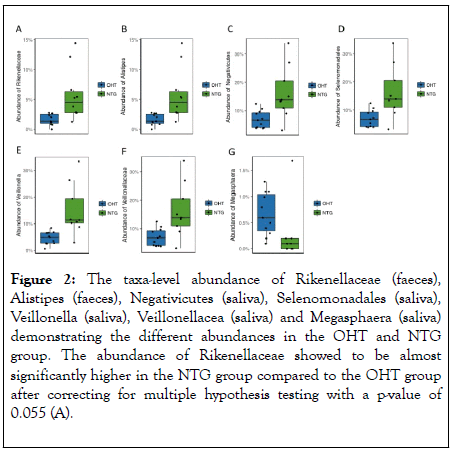
Figure 2: The taxa-level abundance of Rikenellaceae (faeces), Alistipes (faeces), Negativicutes (saliva), Selenomonadales (saliva), Veillonella (saliva), Veillonellacea (saliva) and Megasphaera (saliva) demonstrating the different abundances in the OHT and NTG group. The abundance of Rikenellaceae showed to be almost significantly higher in the NTG group compared to the OHT group after correcting for multiple hypothesis testing with a p-value of 0.055 (A).
An uncoupled microbiome in NTG patients and OHT patients and a coupled microbiome in age- and gendermatched controls
The study demonstrates a correlation between the diversity of bacterial OTUs in a salivary sample and a fecal sample in the same subject in controls only, whereas such correlation between the salivary and fecal samples was not present in the samples from OHT patients and NTG patients. Thus, for controls only, the diversity in the saliva samples is indicative of the diversity of in the fecal samples. For OHT and NTG patients, however, such association is absent.
The UniFrac distance is a measure of dissimilarity in the taxonomic composition between two samples. The UniFrac distance between fecal samples in two controls showed to be significantly associated with the UniFrac distance between the saliva samples from the same two patients (Figure 3).
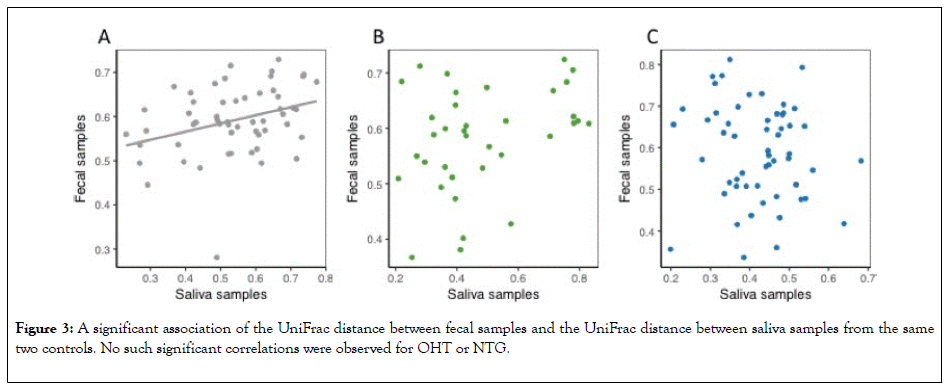
Figure 3: A significant association of the UniFrac distance between fecal samples and the UniFrac distance between saliva samples from the same two controls. No such significant correlations were observed for OHT or NTG.
No such significant association was observed for OHT or NTG. Thus, such significant association among controls equals the association found in the Shannon index (Figure 4).
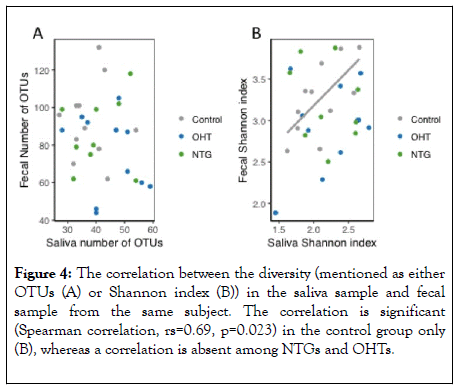
Figure 4: The correlation between the diversity (mentioned as either OTUs (A) or Shannon index (B)) in the saliva sample and fecal sample from the same subject. The correlation is significant (Spearman correlation, rs=0.69, p=0.023) in the control group only (B), whereas a correlation is absent among NTGs and OHTs.
The present study aimed to evaluate microbiomic status of NTG and OHT patients compared to age- and gender-matched control subjects as the human microbiome may play a role in chronic subclinical inflammatory processes. We tested this hypothesis through bacterial 16S analysis of saliva and fecal samples from all subjects. Recent studies have shown that inflammatory processes in the periphery are associated with increased risk of exacerbating neurodegenerative diseases such as Alzheimer ’ s disease and Parkinson ’ s disease [11,29,30]. As the eye is considered an extended part of the brain [21], it is reasonable to believe that similar communication between the gut and the eye is highly relevant in the pathogenesis of neurodegenerative eye conditions.
A recent study has suggested that the oral microbiome is linked to glaucomatous neurodegeneration [22] through a chronic subclinical peripheral inflammation communicated via the guteye axis. Another recent study presents an association between the intestinal microbiomic composition and development of agerelated macular degeneration, indicating that modifications in the intestinal microbiome are linked to the this retinal conditions [31].
The pathogenesis of neurodegenerative retinal diseases of the eye is multifactorial, however it is recognized that disturbed homeostasis caused by low-grade inflammatory processes [5,7,32] plays a significant role. These low-grade inflammatory processes might be affected by peripheral bacterial disturbances as seen with Helicobacter Pylori, which has been associated with glaucoma [33-35]. In line with the impact of the single bacteria species H. pylori, we hypothesized that a mixture of bacteria may also exacerbate glaucoma pathophysiology. In this context, we hypothesized that bacteria might play a more significant role than previously realised in the gut-eye axis through chronic exposure of microbiotic signaling (Figure 5).
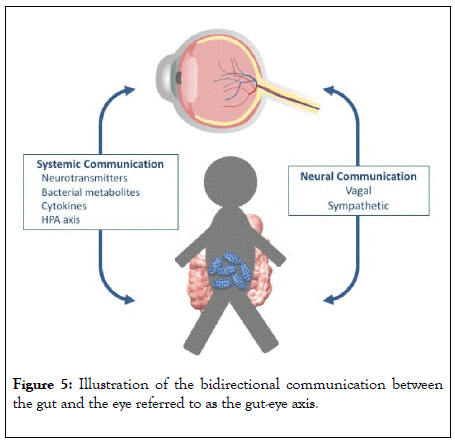
Figure 5: Illustration of the bidirectional communication between the gut and the eye referred to as the gut-eye axis.
The possibility of a peripheral inflammatory impact on the eye has furthermore been confirmed by a recent study demonstrating microglial activation in a mouse model caused by peripheral inflammation [6,36] as well as a novel study that showed increased permeability of the blood-brain barrier by the presence of bacterial pathogens [12]. Such microbiome-bloodbrain barrier communication is highly relevant to possible interpretation of the gut-eye axis.
In this study, we identified an uncoupled association between oral and gut microbiome in NTG patients and OHT patients, whereas we found a significant association between the oral and the gut microbiome in the group of controls. Such uncoupling in the diseased groups may suggest a possible association of an imbalance in the gastrointestinal system for NTG and OHT, however, very limited literature on such possible associations exists. Nonetheless, a recent study hypothesized that the oral and gut microbiome are predictive of each other in healthy adults [37]. Such uncoupling of the salivary and fecal microbiome in the OHT and NTG patients might suggest a common link to retinal neurodegenerative conditions, such as glaucoma.
Our present results indicate that saliva has the potential to be a relevant biomarker in future gut-eye axis studies, since saliva includes more discriminating bacterial abundances on taxa-level compared to faeces. Such a finding suggests that saliva might serve as an optimal microbiome to look for biomarkers in ophthalmic neurodegenerative diseases. That the oral microbiome has been shown to be the least stable over time [37] means that a larger sample or even longitudinal assessments may be necessary to identify whether this apparent association with such a slowly progressive neurodegenerative condition such as glaucoma is indeed real.
The present pilot study has its limitations, especially in the number of participants included in the study to evaluate the association between the microbiomic status in NTG patients and OHT patients compared to controls. Moreover, a large number of NTGs and OHTs are treated with eye drop medication, which may change the composition of conjuntival microflora [38]. If this is possible, eye drop medication may also affect the bacterial composition in salivary samples. Such influence should be taken into consideration in future research, perhaps with sampling of drop-free disease groups such as occurs post-surgery or those with laser trabeculoplasty- controlled IOP, and future studies investigating the gut-eye axis of neurodegenerative retinal diseases will require more participants.
Recent augmented interest in the human gut microbiome has led to increased research in the gut-brain axis. A gut-eye axis might, as the gut-brain axis, include at least three different pathways as shown in figure 1 – a neural pathway, an endocrine pathway and an immunological pathway, where the communication involves both normal, pathogenic and probiotic bacteria [17,39].
The neural pathway involves the vagal nerve, which includes 80 % afferent fibers in direct contact with the layers of the intestinal wall. This innervation is indirectly in contact with the microbiome through diffusion of bacterial compounds and metabolites from the intestinal lumen [39,40]. The endocrine pathway involves the enteroendocrine cells as well as the hypothalamic-pituitary-adrenal-axis (HPA-axis), of which both can be affected by modifications in the human microbiome through changes in neurotransmitter release[17,39]. Imbalances in the endocrine pathway might affect the eye as well as the brain [41]. The mucosal immune system is responsible for the immunological pathway of the gut-brain axis, and certain pathogenic bacteria have shown to induce the production of autoantibodies, and a suggested impact on diseases like Alzheimer’s disease [42] a disease that shares neurodegenerative mechanisms with neurodegenerative retinal diseases [43]. Moreover, the mucosal immune system is assumed to modulate the cytokine level, and hereby regulate the communication between the gut and the brain [44] and potentially the eye as well [5,45].
The human bacterial composition potentially modulates parts of the body situated far away from the intestinal system, and thereby be far more influential on the homeostasis of the human body than previously believed [46]. Therefore, a chronic exposure of microbiotic substances could theoretically have a far more comprehensive impact on the eye than we assume. A secondary neurodegeneration in response to bacterial composition might exacerbate inner retinal disease pathology. If such hypothesis is confirmed by future research, it will be of significant impact on therapeutic strategies.
In summary, the current study did not find any different microbiome signatures in between patients with NTG and OHT. Thus, the apparent differences in retinal ganglion cell resistance within the two groups did not seem to be related to the microbiome. In contrast, the fecal and saliva microbiome of NTG and OHT patients were identical, but different from the age- and gender-matched controls. In this context, the oral and gut microbiome were linked in the control subjects, but relatively unlinked in both patients with NTG and OHT. Over all, the study could not confirm a direct association between glaucomatous retinal ganglion cell loss and the microbiome. We suggest that the microbiome is similar in patients with NTG and OHT, but different in both from age- and gender-matched controls. Future and larger studies are needed to clarify the final role of the microbiome in glaucoma.
The authors of the present study are very grateful to the participants of the study and to their relatives. Furthermore, the authors are thankful to the financial support of the present study by A.P. Møller Foundation, Fight for Sight (Denmark) and Synoptik- fonden.
Citation: Toft-Kehler AK, Vibæk J, Kolko M, Gazzard G (2020) Investigation of the Association between the Oral and the Gut Microbiome in Glaucoma. J Clin Exp Ophthalmol. 11:826. DOI: 10.35248/ 2155-9570.20.11.826
Received: 26-Feb-2020 Accepted: 11-Mar-2020 Published: 18-Mar-2020 , DOI: 10.35248/2155-9570.20.11.826
Copyright: © 2020 Toft-Kehler A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.