Journal of Glycomics & Lipidomics
Open Access
ISSN: 2153-0637
ISSN: 2153-0637
Review Article - (2015) Volume 5, Issue 1
The role of lipids in human health and disease is taking the center stage. In the last decades, there has been an intense effort to develop suitable methodologies to discover, identify, and quantitatively monitor lipids in biological systems. Recent advancement of mass spectrometry technology has provided a variety of tools for global study of the lipid “Omes”, including the quantification of known lipid molecular species and the identification of novel lipids that possess pathophysiological functions. Lipidomics has thus emerged as a discipline for comprehensively illuminating lipids, lipidderived mediators and lipid networks in body fluids, tissues and cells. However, owing to the complexity and diversity of the lipidome, lipid research is challenging. Here, the experimental strategies for lipid isolation and characterization will be presented, especially for those who are new to the field of lipid research. Because lipids are known to participate in a host of protein signaling and trafficking pathways, the review emphasizes the understanding of interactions between cellular components, in particular the lipid-protein interrelationships. Novel tools for probing lipid-protein interactions by advanced mass spectrometric techniques will be discussed. It is expected that by integrating the approaches of lipidomics, transcriptomics and proteomics, a clear understanding of the complex functions of lipids will eventually be translated into human diseases.
<Keywords: Lipids, Lipidomics, Lipid-protein interactions, lipoproteomics
The main differences between lipids and other biomolecules (carbohydrates, proteins and nucleic acids) are their solubility in non-polar solvents and structure of long hydrocarbon chains. Based on the physical and chemical properties, eukaryotic and prokaryotic lipids are grouped under eight categories, each of which contains distinct classes, subclasses, subgroups and subsets of lipid molecules (Table 1) [1]. When studying lipids from a system scale, one of the key challenges is to address the lipid functionality at many physiological levels from metabolism, signaling pathways to spatial regulation, as well as the interactions with other “Omics”. Lipidomics is to map the entire spectrum of cellular lipids in biological systems, including metabolic pathways, lipid–lipid and lipid-protein interactions [2]. It complements proteomics, genomics and metabolomics for providing a more comprehensive understanding of system biology in health and disease [3].
| Category | Core structure | Classes/Subclasses |
|---|---|---|
| Fatty acyls | 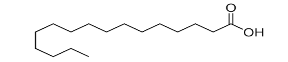 |
Fatty acids and conjugates, Fatty esters, fatty alcohols, Fatty amides, eicosanoids |
| Glycerolipids | 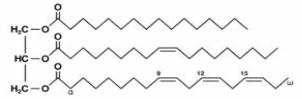 |
Monoacylglycerols, Monogalactosyldiacylglycerols, Diacylglycerols, Digalactosyldiacylglycerols, Triacylglycerols |
| Glycerophospholipids | 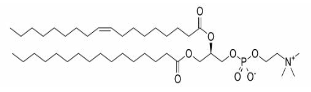 |
Phosphatidic acids, Phosphatidylethanolamine, Phosphatidylcholine, Phosphatidylglycerols, Phosphatidylserines, Phosphatidylinositols, Cardiolipins |
| Sphingolipids | 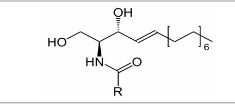 |
Ceramides, sphingosines, Sphingosine-1-phosphates, Gangliosidemannoside 3, Sphingomyelins Sulfatides |
| Sterol lipids |  |
Cholesterols, Cholesteryl esters, Cholesteryl sulfates, Steroids, Bile acids |
| Prenol lipids |  |
Isoprenoids, Polyprenoids, Quinines, Hydroquinines, |
| Saccharolipids |  |
Acylaminosugars, Acylaminosugarglycans, Acyltrehaloses, Acyltrehalosesglycans |
| Polyketides |  |
Macrolide polyketides, Aromatic polyketides, Non-ribosomal peptide/polyketide hybrids |
Table 1: Categories, classes and subclasses of lipids.
As lipids are not genetically encoded, the total number of distinct chemical entities in lipidome is poorly defined. In general, lipids are derived from the condensation of two distinct types of building blocks, ketoacyl or isoprene subunits [4,5]. However, the biosynthetic and metabolic pathways of lipids are enormously diversified. Due to the combinatorial nature of lipid biosynthesis, the various backbone, headgroup and acyl chains can give rise to thousands of lipid species. The dynamic nature of lipidomics requires not only better analytical techniques, but also tactical sample processing and integrative computational tools. Due largely to the advanced technology development, lipidomics is now opening a novel avenue in the discovery of biomarkers for predictive and preventive medicine. This review focuses on the strategies to study lipids in health and disease, with specific emphasis on lipid-protein interactions.
As lipids are not genetically encoded, the total number of distinct chemical entities in lipidome is poorly defined. In general, lipids are derived from the condensation of two distinct types of building blocks, ketoacyl or isoprene subunits [4,5]. However, the biosynthetic and metabolic pathways of lipids are enormously diversified. Due to the combinatorial nature of lipid biosynthesis, the various backbone, headgroup and acyl chains can give rise to thousands of lipid species. The dynamic nature of lipidomics requires not only better analytical techniques, but also tactical sample processing and integrative computational tools. Due largely to the advanced technology development, lipidomics is now opening a novel avenue in the discovery of biomarkers for predictive and preventive medicine. This review focuses on the strategies to study lipids in health and disease, with specific emphasis on lipid-protein interactions.
Lipids are a diverse group of hydrophobic or amphipathic small molecules that possess many functions, including the maintenance of membrane structure, energy storage, signal transduction and regulation of gene expression (Hyotylainen and Oresic, 2014). Lipids are generated and metabolized by enzymes. The metabolism of lipids is regulated by a coordinated network of signaling pathways that are dynamically regulated by the environment. Virtually all lipid pathways are interconnected and perturbations in one branch usually will affect the entire lipid metabolic network (i.e. resulting in global lipidome changes). Thus, understanding the anabolism and catabolism of lipids is essential for research and development in the field of lipidomics. Furthermore, altered lipid metabolism plays important roles in the pathogenesis of most of the common diseases, such as type 2 diabetes, cancers, coronary artery and neurodegenerative diseases. Here, the major classes of lipids, including glycerolipids, glycerophospholipids, sphingolipids and sterol lipids, will be discussed.
Fatty acids and their derivatives
Fatty acids are aliphatic monocarboxylic acids with a large diversity in structure ranging from simple saturated carbon chains to unsaturated, branched, cyclic and cis/trans configurations. According to the length of carbon atoms, they are classified as short (less than 6 carbons), medium (from 6 to 12 carbons), long (from 13 to 21 carbons) and very long (more than 22 carbons) chain fatty acids. In addition, functional groups including keto, hydroxyl, peroxy and epoxy groups can be attached to fatty acids. The majority of fatty acids exist in the form of esters and amides. Free fatty acids constitute only a small portion of the total fatty acids [6].
Fatty acids are stored primarily as triacylglycerols and sterol esters [7]. Because of the highly reduced chemical structure, fatty acids yield more than twice as much energy upon oxidation compared with polysaccharides, which allows fat to be the most efficient form for storing excess energy. On demand, fatty acids are released from storage or membrane lipids by hydrolysis mediated by lipases. Fatty acids are synthesized de novo from acetyl-CoA by fatty acid synthase (FAS) to yield palmitate (16:0), which then be either desaturated to palmitoleate (16:1) by stearoyl-CoA desaturase (SCD) or elongated by an elongase (ELOVL6) to stearate (18:0) [8]. Stearate can be further desaturated to oleate (18:1) by SCD [9]. Eukaryotes lack the enzymatic system to introduce double bonds at position omega (ω)-6 or lower. Thus, these subtypes of fatty acids or their precursors are obtained from the diet. In human, linoleic acid (18:2, ω-6) and alpha-linolenic acid (18:3, ω-3) are essential fatty acids and form the parent molecules for arachidonic (AA, 20:4) and eicosapentaenoic acid (EPA, 20:5)/ docosahexaenoic acid (DHA, 22:6), respectively. AA, EPA and DHA, are commonly found at the sn-2 position of glycerophospholipids for the synthesis of classical (prostaglandins leukotrienes, thromboxanes, lipoxins) and nonclassical (endocannabinoids, neuroprotectins and resolvins) eicosanoids [10]. In mammals, fatty acid synthesis and metabolism are tightly controlled by the nuclear transcription factor sterol regulatory element binding proteins (SREBP), which regulate genes such as FAS, SCD and ELOVL6 [11].
Historically, fatty acids were considered to be simple membrane components serving as the basic building blocks for various complex lipids, such as glycerolipids and glycerophospholipids [12]. It is increasingly recognized that fatty acids are important signaling molecules regulating cellular homeostasis and functions [13]. As the integral components of cellular membranes, fatty acid composition determines the fluidity and the movement of molecules within and across the membrane, the activity of membrane proteins and receptors, the arrangement of membrane microdomains, the trafficking and signal transduction processes [14]. With the help of transporting proteins, fatty acids are distributed via blood circulation to organs and act as ligands of G protein-coupled receptors (e.g. GPR41 and GPR43) and nuclear receptors (e.g. PPARα), which play crucial roles in the regulation of energy expenditure [15]. Aberrant fatty acid metabolism causes pathological consequences, including insulin resistance, diabetes, cancer, fatty liver and cardiovascular diseases [16-18].
Glycerophospholipids
Lipids with a phosphate functional group are classified as phospholipids, which account for ~60% lipid mass in eukaryotic cells [19]. Phospholipids, including glycerophospholipids and phosphosphingolipids, are the primary building blocks of biological membranes and the precursors for signaling molecules such as inositol triphosphate and diacylglycerol, which are produced by various phospholipases [20]. Phospholipids are also sources of lipid mediators, including prostaglandins and leukotrienes that regulate cell homeostasis and inflammation [21,22]. Changes in profile and concentration of phospholipids can result in major consequences on cell function and viability. For example, translocation of the phosphatidylserine head group to the outer leaflet of the plasma membrane is involved in cellular apoptosis and the recognition by macrophages for subsequent phagocytosis [23-31]. Drugs-induced phospholipidosis is one of the major concerns in drug development and clinical treatment [24].
Glycerophospholipids consist of a glycerol backbone with a hydrophilic head group at sn-3 position and different combinations of fatty acids at sn-1 and sn-2 positions. Saturated or monounsaturated fatty acids are mainly found at sn-1 position, whereas polyunsaturated fatty acids conjugated at sn-2 position. The polar head groups define the subclasses of glycerophospholipids (Table 1). During glycerophospholipids synthesis, the first step is the transfer of a fatty acid to glycerol-3-phosphate to form lysophosphatidic acid (LPA), which is further acylated to phosphatidic acid (PA). After conversion to CDPdiacylglycerol, PA provides the precursor for phosphatidylinositol and phosphatidylglycerol. Alternatively, PA is dephosphorylated to generate diacylglycerol, which fuels the synthesis of phosphatidylcholine, phosphatidylethanolamine and phosphatidylserine. Phosphorylated choline/ethanolamine are converted to CDP-choline/ethanolamine, which are then used for head groups of phosphatidylcholine and phosphatidylethanolamine, respectively (the so-called Kennedy pathway). Both phosphatidylcholine and phosphatidylethanolamine are substrates for phosphatidylserine synthesis, which may be converted to phosphatidylethanolamine by decarboxylation in the mitochondria. Thus, the metabolism of phosphatidylethanolamine and phosphatidylserine is tightly linked. Phosphatidylethanolamine can be derived additionally from N-methylation of phosphatidylcholine (Figure 1). These pathways generate glycerophospholipids with different profiles of fatty acid tails. The CDP-choline pathway produces those with medium, saturated chain species (e.g. 16:0/18:0), whereas the methylation pathway results in more diversified long chain and polyunsaturated species (e.g. 18:0/20:4) [25]. Disruption of most phospholipid biosynthetic pathways leads to embryotic lethality in mice (Vance and Vance, 2009). The newly synthesized glycerophospholipids are dynamically modified by acyl chain remodeling and translocated between membranes of different subcellular organelles [26]. In addition, base-exchange represents a minor pathway for synthesis of most phospholipids to provide a rapid energy-independent means of replenishment (Vance and Vance, 2004).
Figure 1: Synthesis of Glycerophospholipids. Glycerophospholipids are shown in blue; substrates and by-products for glycerophospholipid metabolism are shown in black; and all important enzymes are shown in orange. R and R’ in molecule structure represents all possible functional groups derived from fatty acid. Abbreviations: CDP, cytidine diphosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; PgpP, phophatidylglycerolphophate phosphatase; PgpS, phophatidylglycerolphophate synthase; PtdsD, phosphatidylserine decarboxylase; PtdsS1, phosphatidylserine synthase-1; PtdsS2, phosphatidylserine synthase-2
The composition of glycerophospholipids differs from one organelle to another. Phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol and cardiolipin are the major lipid constituents of the mitochondrial membranes, rendering the surface negatively charged. Phosphatidylcholine and phosphatidylinositol are bilayer lipids with cylindrical molecular shapes, i.e. the cross-sectional areas of the lipid head group and the acyl chains are similar. As a result these lipids self-assemble into bilayers upon hydration. In phosphatidylethanolamine, the cross-sectional area occupied by the head group is smaller than that occupied by the acyl chains, resulting in a conical molecular shape, and tend to adopt non-bilayer structure [27].
Sphingolipids
Sphingolipids consist of a sphingoid backbone that is N-acylated with various fatty acids to form ceramide species with hundreds of distinct head groups. These fatty amino alcohols, including ceramide (N-acyl-sphingosine), sphingomyelin and different glycosphingolipids, have been implicated in the pathogenesis of metabolic disorders, cancer and neurological diseases [28]. The sphingolipids in yeast, mammals and insects have very different head group decorations, hydroxylation patterns and lengths of fatty acids chains.
The synthesis of sphingolipids starts in the endoplasmic reticulum (ER) from the condensation of serine and palmitoyl coenzyme A (CoA) to form 3-ketosphinganine, which is catalyzed by serine palmitoyltransferase (Figure 2). The condensation product is reduced to dihydrosphingosine (sphinganine), which is then N-acylated by one of six ceramide synthases (CerS1-CerS6) with specific acyl chains (typically with saturated or mono-unsaturated fatty acids with 14 to 26 carbons). Dihydroceramides are subsequently dehydrogenated to ceramides. Metabolism of ceramide to sphingomyelin and glycosphingolipids mainly occurs in the Golgi. Ceramide transfer protein is responsible for transportation of ceramides from ER to Golgi during sphingomyelin synthesis, whereas vesicular transport is involved in glucosylceramide synthesis. Ceramides can also be phosphorylated in the Golgi by ceramide kinase to form ceramide-1-phosphate (C1P).
Figure 2: Sphingolipid metabolisms. Sphingolipids are shown in blue; substrates and by-products for Sphingolipids metabolism are shown in black; and all important enzymes are shown in orange. Biochemical reactions are represented by solid arrows, and inter-organelle transportations are represented with dashed arrows. R and R’ in represent possible functional groups derived from fatty acid. Abbreviations: ALDH3A2, fatty aldehyde dehydrogenase; C1P, ceramide-1-phosphate; ER endothelium reticulum; Pm-CoA, palmitoyl coenzyme A; S1P, sphingosine-1-phosphate; and TGN, trans-Golgi network.
Sphingomyelin is synthesized by the transfer of phosphocholine from phosphatidylcholine to ceramide with the generation of diacylglycerol, a pathway catalyzed by sphingomyelin synthase and representing an interconnection between sphingolipid and glycerolipid metabolism. Sphingomyelin is the major component found in the membranous myelin sheath that surrounds nerve cell axons [29]. The synthesis of glycosphingolipids is initiated by the addition of glucose to ceramide by glycosylceramide synthase and requires the transfer of glucosylceramide to the trans-Golgi network by four-phosphate adaptor protein 2, which also regulates vesicular trafficking from the Golgi to the plasma membrane [30].
Sphingolipids are degraded in lysosomes by removing the head groups to form ceramides. Sphingomyelinase hydrolyzes sphingomyelin and releases ceramide, which may be further degraded by ceramidase to sphingosine. Sphingosine can be recycled by reacylation back to ceramide, thus fueling the salvage pathway. This reutilization pathway plays a significant role in sphingolipid homeostasis. Sphingosine can also be phosphorylated by sphingosine kinase to form sphingosine- 1-phosphate (S1P), a bioactive metabolite and a key intermediate in the sphingolipid-to-glycerolipid metabolic pathway. Irreversible degradation of S1P by S1P lyase generates phosphoethanolamine and hexadecenal, another key intermediate in the sphingolipid-to-glycerolipid metabolic pathway [31]. The fatty aldehyde dehydrogenase ALDH3A2 converts hexadecenal to hexadecenoate, which is utilized for the formation of palmitoyl-CoA in glycerolipid synthesis. The sphingolipid metabolites, including ceramide, C1P and S1P are important signaling molecules that regulate cell growth, survival, trafficking and integrity.
Sterol lipids
Sterols are constituents of the cellular membranes (myelin in the nervous system) and precursors of steroid hormones and bile acids. In mammals, cholesterol is the main sterol. Other sterols, including ergosterol and 24-methyl sterols, are absent from mammalian cells but required for parasitic growth and viability [32]. Cholesterol homeostasis is regulated by both endogenous and exogenous pathways of cholesterol metabolism. Endogenous cholesterol is synthesized by the liver (~10%) and extrahepatic tissues, enters the circulation as a component of lipoproteins, and secreted into bile. The brain contains more cholesterol than any other organs, and brain cholesterol is almost all synthesized locally. Exogenous cholesterol is absorbed in the intestine and ultimately enters the circulation as a component of chylomicrons. Circulating cholesterol concentrations are maintained by biosynthesis as well as absorption. The biochemical pathway of cholesterol synthesis is schematically presented in Figure 3. Cholesterol synthesis occurs in the cytoplasm and microsomes. It starts from the reduction of acetyl-CoA to generate 3-hydroxy-3-methylglutaryl-CoA (HMGCoA), which then is converted to mevalonate, a process catalyzed by HMG-CoA reductase and using NADPH as a cofactor. Mevalonate is then activated by two successive phosphorylations, followed by an ATP-dependent decarboxylation to yield an activated isoprenoid molecule, isopentenyl pyrophosphate (IPP). The subsequent multiple steps of IPP condensation produce squalene. A two-step cyclization of squalene produces lanosterol. Through a series of additional reactions catalyzed by 7-dehydrocholesterol reductase, lanosterol is converted to cholesterol. The post-lanosterol steps of cholesterol biosynthesis are divided into Bloch and Kandutsch–Russell pathways, which share enzymatic stages but produce C24 double-bond reduced cholesterol at different steps. HMG-CoA formation is the rate-limiting step in sterol synthesis that is controlled by cholesterol levels. If cholesterol uptake is low, liver and small intestine will synthesize cholesterol accordingly. If HMG-CoA is not needed, such as when intake of dietary cholesterol is high, it is degraded quickly to acetoacetate and acetyl-CoA by HMG-CoA lyase in the mitochondrial matrix compartment. Steroid hormones are generated in specialized organs, such as adrenal glands and gonads, each expressing a different set of steroidogenic enzymes. All steroid hormones are derived from a single precursor, cholesterol, which is transformed to the steroid pregnenolone in the mitochondrial matrix by the enzyme CYP11A1. Translocation of cholesterol to mitochondria is a key step in steroidogenesis that is accomplished by means of transport proteins [33].
In the intestinal lumen, the cholesterol esters are cleaved by pancreatic cholesteryl ester hydrolase, which is produced by the exocrine pancreas. Free cholesterols, along with other lipids and fat-soluble vitamins, are solubilized into micelles and absorbed by enterocytes. After absorption, the free cholesterol is re-esterified to cholesteryl ester by acyl CoA:cholesterol acyltransferase and packaged with other lipids into chylomicrons, which are secreted into the mesenteric lymph and ultimately into plasma [34]. Once in circulation, chylomicrons are hydrolyzed by lipoprotein lipase at the endothelial surface of vessels and reduced to chylomicron remnants, which can then be removed from the circulation by the liver or, if they are small enough, may be able to penetrate the endothelial surface of the arterial wall, where they may contribute to plaque formation [35]. Abnormal deposition of cholesterol and cholesterol-rich lipoproteins in the arteries is of particular importance clinically, as such deposition eventually leading to the development of atherosclerosis. Elevated levels of cholesterol in the liver lead to an increased production of verylow- density lipoprotein (VLDL) and/or low-density lipoprotein (LDL) particles, as well as down-regulation of the LDL receptor. Cholesterol is acylated before incorporated into VLDL and LDL. VLDLs that contain both triglycerides and cholesterol are transported to target organs (e.g. adipose tissue and muscle) to unload triglycerides. The remaining cholesterol rich LDLs are recognized by other cell types and taken into the cells. After endocytosis via LDL receptor, cholesterol is extracted within the lysosomal compartment for membrane biosynthesis. The LDL receptor recycles to the cell surface.
Cholesterol overload is prevented by its intracellular esterification and subsequent storage in lipid droplets, and by its release. The relative contribution of each pathway to cholesterol homeostasis is cell type-specific. Cholesterol is released either as a complex with apolipoprotein-containing lipoproteins via members of the ATPbinding cassette transporters or after conversion to oxysterols, a broad class of cholesterol derivatives that includes enzymatically derived metabolites and compounds generated by non-enzymatic reactions [36]. In brain cells, especially neurons, the only route to eliminate excess cholesterol is converting it to 24S-hydroxycholesterol by the enzyme cholesterol 24-hydroxylase, Cyp46a1. High levels of 24(S),25- epoxycholesterol, a metabolite generated via blocking the mevalonate pathway, have been associated with several neurodegenerative diseases, implying that these metabolites play crucial signaling functions [37]. The efflux of cholesterol and phospholipid from peripheral cells is induced by high-density lipoprotein (HDL) and apolipoprotein A-I (apoA-I). Nascent HDL takes up cholesterol from cell membranes and other lipoproteins, a process of major biological importance in the prevention of cardiovascular disease. Extracellular esterification of free cholesterol by lecithin:cholesterol acyltransferase (LCAT) provides the driving force for cellular cholesterol removal, which is facilitated by ATP binding cassette transporter G1/G4 (ABCG1/ABCG4) and ABCA1, and involves the lipidation of lipid-free or lipid-poor apoA-I with cellular cholesterol and phospholipids [38]. In situations of excess cellular cholesterol, the nuclear liver X receptors induce the transcription of ABCA1 and ABCG1 and thus cholesterol efflux. Significant proportions of HDL-cholesterol are removed by selective uptake through SR-BI into the liver and steroidogenic organs [39].
Distribution of lipids
In circulation, the abnormal distribution of lipoprotein particles is closely associated with various disease conditions. VLDL, LDL and HDL display characteristic patterns of lipid class and species. Phospholipids are unequally distributed across lipoproteins. HDL represents a major carrier of phosphatidylcholine, lysophosphatidylcholine, phosphatidylethanolamine and its derivative plasmalogen, containing over 50% of these lipid classes present in human serum [40]. Phosphatidylcholine accounts for 33-45% of total lipids in HDL [41]. Comparing to other lipoproteins, HDL is enriched in phosphatidylcholine containing polyunsaturated fatty acids. Lysophosphatidylcholine, a product of the phosphatidylcholine hydrolysis, constitutes up to 15% of HDL but only 2-3% in apoBcontaining lipoproteins. The 16:0 lysophosphatidylcholine is enriched in HDL, whereas the 18:0 species is predominantly associated with VLDL and LDL. For plasmalogen, a phospholipid with antioxidative properties, species containing arachidonic acid residues predominate in HDL, whereas species containing 18:1 and 18:2 residues are depleted. The plasma phosphatidylethanolamines are mainly the 36:2 and 38:4 species, which are evenly distributed across VLDL, LDL and HDL. All other negatively charged phospholipids, including phosphatidylinositol, phosphatidylserine, phosphatidylglycerol and phosphatidic acid are present in lipoproteins in low amounts. Compared to LDL, HDL is depleted in glycosphingolipids and gangliosides. Sphingomyelin accounts for 5-10% of total lipid in HDL, mainly the d18:1/16:0 and d18:2/24:0 species. By contrast, LDL contains the largest amount of sphingomyelin, accounting for over 50% of plasma sphingomyelin. Similarly, ceramides are preferentially carried by LDL (60% of total plasma ceramides) compared with HDL (25%) and VLDL (15%) [40]. An increased sphingomyelin to phosphatidylcholine ratio enhances the susceptibility of LDL to secretory sphingomyelinase, which leads to ceramide generation and the formation of aggregated LDL with a high atherogenic potential [42].
The ratio of shell to core lipids and the composition of the amphipathic and neutral lipids are different in the subclasses of HDL particles [41]. Steroids in HDL particles are dominated by cholesterol (5-10% of lipid) and located in the surface lipid monolayer. LCAT catalyzes the trans-esterification between phospholipids and cholesterol to produce cholesteryl esters (30-40% of HDL lipids). This reaction displaces the cholesterol moiety from the surface lipid monolayer into the lipid core of HDL. Cholesteryl esters in HDL are present mainly in the form of cholesteryl linoleate [41]. The cholesteryl esters can be transferred to apoB-containing lipoproteins in exchange for triglycerides, which are located in the HDL lipid core and contain mainly species of oleic, palmitic and linoleic acid moieties. The shells of large HDL particles are enriched with sphingomyelins and ceramides, whereas those of small particles contain more phosphatidylserine, phosphatidic acid and phosphatidylcholine. The shells of very small particles are enriched in lysophosphatidylcholine and depleted of free cholesterol [43]. The side chain distribution is shifted towards more unsaturated moieties in smaller particles comparing to larger particles. These changes influence the physical properties such as surface charge and membrane fluidity of HDL particles. The smaller HDL particles have a higher capability to stimulate cholesterol efflux and protect the oxidation and pro-thrombotic activity of LDL.
In mammalian cells, lipids are distributed within membranes of multiple compartments that display distinct composition of both lipids and their interacting proteins. The membrane lipid composition is different between cell types and subcellular compartments. Lipid composition of subcellular organelles not only influences their membrane structure and function, but also serves as a descriptor of an organelle’s identity. For instance, the plasma membrane is enriched with phosphatidylinositol 4,5-biphosphate (PI(4,5)P2), whereas phosphatidylinositol 4-phosphate (PI(4)P) is the predominant phosphoinositide in Golgi. Consequently, the protein compositions of these two membrane compartments vary significantly, giving rise to distinct morphologies and functional properties [44]. Lipids are symmetrically distributed between the two leaflets of the membrane bilayer in endoplasmic reticulum. On the other hand, the Golgi, endosomal and plasma membrane display an asymmetric lipid distribution, with phosphatidylcholine, sphingomyelin, and glycosphingolipids enriched at the luminal side, and phosphatidylethanolamine, as well as negatively charged phospholipids, including phosphatidylinositol and phosphatidylserine, enriched at the cytosolic leaflet [45]. Mitochondria contain a complex membrane system and the membrane lipid composition depends on the interplay with the endoplasmic reticulum, from which some of the lipids are imported. The mitochondrial contents of specific lipids, such as cardiolipins, can be modulated by dietary intervention, genetic manipulation and disease conditions [46].
In summary, to understand the dynamic functions of lipids, knowledge about the total cellular lipidome alone is not sufficient, as it does not provide information about the spatial distribution of the lipids within cells. The combined information of the localization and the composition of subcellular lipids is a starting point for the molecular elucidation of the lipid networks. Moreover, environmental and pathophysiological conditions are to be considered when analyzing the dynamic changes of the intracellular and extracellular lipidomes.
Lipid-protein interactions
Lipids and their metabolites work together with proteins to regulate many cellular functions and signal transduction pathways. Lipid interactions determine the localization, structure and function of proteins [47]. The highly specific interactions between lipids and proteins provide a promising area for the discovery and development of therapeutic targets [48]. For instance, phosphoinositides are an important class of cellular signaling lipids produced by phosphatidylinositol phosphate kinases and act on a large number of protein effectors [49,50]. Palmitoylation and myristoylation play an important role in the trafficking, compartmentalization and membrane tethering of proteins [51,52]. The specificity of lipid-enzyme interactions determined by the geometry of the catalytic activity is critical for the enzyme cyclooxygenase-2 (COX-2) to produce lipid mediators with opposing (pro- and anti-inflammatory) functions from chemically distinct glycerophospholipids substrates with ω-6 and ω-3 fatty acyl, respectively [53]. Thus, understanding lipid–protein interactions has important biological or medical implications.
The unique physico-chemical property shared by all lipid species, i.e., their hydrophobicity, is accompanied by a poor solubility in an aqueous environment. Thus, specific proteins which can reversibly and non-covalently associate with lipids, designated as lipid binding proteins or lipid chaperones, are needed for the transportation of lipids in compartments such as blood plasma and the cellular soluble cytoplasm. These lipid binding proteins determine the bio-availability of their ligands, and thereby markedly influence the subsequent processing, utilization, or signaling effect of lipids [54]. Depending on the strength of their interactions with the protein and the consequent rate of exchange, lipids have been referred to as different terms. The bulk membrane lipids exchange very rapidly, and interact nonspecifically with the protein through the physical properties. The shell of lipids at the surface of the protein is usually referred to as annular lipids. Other lipids buried within a membrane protein, in deep grooves at the protein surface, between transmembrane α-helices, or at the protein–protein interface are referred to as non-annular lipids. The bindings of non-annular lipids at the interface between transmembrane helix bundles can be modulated by hydrophobic inhibitors, such as the small molecules that occupy the lipid binding cavities of Ca2+-ATPase [55]. The binding of peripheral membrane proteins to anionic or amphipathic lipids constitutes another class of specific lipid-protein interactions that are often involving lipid-binding domains, such as PH, FYVE, PX, C2 and ENTH, via electrostatic forces [56]. These interactions are common in cell signaling and membrane trafficking.
In circulation, lipids are transported by lipoproteins. The various lipid species are not only important structural components, but also modulate the function of lipoproteins. For example, the antiatherogenic potential of HDL can be enhanced by phosphatidylcholine and sphingomyelin, whereas adding sphingomyelin in reconstituted HDL inhibits LCAT activity [57]. HDL particles are smaller and richer in protein contents when compared to other plasma lipoprotein classes. Levels of HDL are inversely associated with the risk of coronary artery disease and its thrombotic complications [39]. Thus, it has been assumed that raising HDL-cholesterol levels reduce cardiovascular mortality. Unexpectedly, pharmacologic interventions to increase HDL-cholesterol did not translate into the reduction of cardiovascular risks [39]. Recent studies reveal that HDL composition, rather than the circulating levels, determines its functional properties. Approximately half of the total HDL masses are lipids, consisting of phospholipids and sphingomyelin (40-60%), cholesteryl esters (30-40%), triglycerides (5- 12%) and free cholesterol (5-10%). More than 200 individual molecular lipid species have been identified in HDL particles (Kontush et al., 2013). In humans, cholesteryl-ester transfer protein (CETP) transfers HDL-cholesteryl esters to apolipoprotein B-containing lipoproteins, which then are removed by the LDL receptor pathway or the liverindependent transintestinal excretion of cholesterol [39]. Under pathological conditions, such as inflammation, both the proteome and lipidome of HDL are significantly altered [58].
Membrane proteins associate with lipids through multiple mechanisms, including unspecific hydrophobic association and electrostatic interactions, as well as specific covalent bonding [59]. The mechanisms by which a protein interacts with lipids determine the specificity and affinity of membrane binding, the morphology and dynamics of the membrane. The lipid distribution and composition in membranes affect protein localization. For example, if the sterol concentration were equalized, mitochondrial tail-anchored proteins would mistakenly localize to the endoplasmic reticulum of yeast cells [60]. Fluxes of cargoes at the Golgi depend on distinct membrane structures. There is a gradient of glycerophospholipids across cis-to-trans cisternae of the Golgi. Reducing the lipid order of the Golgi membrane impairs vesicle fission and cargo export in animal cells [61]. In yeast, sphingolipid-enriched and sterol-enriched domains are segregating at post-Golgi compartments and involved in protein sorting. The lipids and proteins in cell membranes are able to laterally segregate via lipid rafts [the nanoscale assemblies containing sphingolipid, cholesterol and specific proteins] that play important roles in signal transduction and vesicle transportation [62]. The dynamic liquid-liquid immiscibility and membrane bioactivity is created by sphingolipid-cholesterol selfassembly with proteins in lipid rafts [63]. Decoration of proteins with glycosylphosphatidylinositols are critical for their trafficking to lipid rafts and the outer leaflet of the plasma membrane [64]. Perturbation of membrane microstructures by HDL may explain the observed antiinflammatory effects in innate immune cells. In macrophages, HDLinduced cholesterol removal disrupts the formation of cholesterol enriched plasma membrane domains (e.g., lipid rafts), leading to the inhibition of foam cell formation and inflammation [39].
In mitochondria, proteins involved in fusion, division and degradation bind to phospholipids and the interactions generate a wide array of mitochondrial responses [65]. Cardiolipins are phospholipids located at mitochondrial inner membrane and critically involved in regulating mitochondrial structure, oxidative phosphorylation and biogenesis [66]. Structurally, cardiolipin consists of two phosphatidyl residues linked by a glycerol moiety and attached with a total of four fatty acyl chains [67]. Cardiolipin binding is essential for the stability of respiratory chain supercomplexes and oxidative generation of ATP [68]. In mammalian hearts, cardiolipins are enriched with symmetric linoleic acid, which is essential for the high affinity binding of these lipids to membrane proteins and maintaining mitochondrial respiration. Replacement of 18-carbon unsaturated fatty acids with those of 22-carbon species is found in myocardial cardiolipins of diabetic animals [69]. In fact, the loss of linoleic acid content due to cardiolipin remodeling occurs in various cardiac disorders, including ischemic/reperfusion injury, heart failure, diabetic cardiomyopathy and aging-induced cardiac dysfunction [70-72]. Cardiolipins are bilayer preferring lipids that upon charge neutralization, acquire non-bilayer propensity that confers negative curvature stress to membranes, which is important for processes such as fusion and fission via interacting with dynamin-related protein, Mgm1 [65].
The wide range of lipid building blocks produces a large array of combinations, especially for sphingolipids and glycerophospholipids. Theoretically, rearranging the common eukaryotic lipid motifs (i.e. without the consideration of isomeric lipids that differ only in doublebond position, backbone substitution or stereochemistry) can give rise to more than 180,000 phospholipid structures in a given cell. The structural diversity of lipids presents considerable challenges to comprehensive lipid analysis. For instance, it is almost impossible to collect data for all classes of lipids in most biological samples using a single method for extraction, chromatography separation and analytical detection.
Lipidomics is to comprehensively elucidate lipid-based information, including the identification and quantification of lipids and derivatives, and to establish pathways and networks of lipid species in biological systems [73]. Increased throughput, higher sensitivity and higher chromatographic resolution remain the key goals for lipidomics research. The LIPID MAPS Consortium (www.lipidmaps.org) has established detailed procedures for sample extraction, separation and quantitative analysis for major lipid categories [4]. The structural and quantitative analysis of lipids are mainly performed by mass spectrometry (MS), with which lipid ions are identified by their massto- charge ratio (m/z). The peak intensity corresponds to the abundance of a lipid molecule. Normalization of the intensity to the standards enables absolute quantification of the lipid molecules. Fragmentation of the lipids specifies their molecular identities for unique lipid species and subspecies
Extraction of Lipids
Lipids are embedded in complex matrixes. Prior to analysis, isolation and/or fractionation are necessary in order to remove any non-lipid components, such as proteins, saccharides and other small molecules. Generally, two extraction methods are used to separate lipids from biological samples. Liquid–liquid extraction allows an instant partition of the lipids. The Folch method was originally designed to use chloroform/methanol solvent with or without incorporated H2O as the extraction matrix [74]. The high efficiency of chloroform/methanol/ H2O extraction method is due to the capability to penetrate through cell membrane, higher polarity and stronger interaction with hydrogen bond. The proportions of chloroform, methanol and H2O depend on the moisture content of the samples. Other liquid-liquid extraction methods include the use of hexane/isopropanol solvent [75], the one phase butanol/methanol mixture combined with two phases heptane/ ethyl acetate extraction matrix, a methyl tert-butyl ether-based solvent [76], a combined extraction with dichloromethane and a methyl tertbutyl ether/hexafluoroisopropanol mixture [77], and so on.
Solid-phase extraction does not require the partition of lipids in solvent/water mixture, but use stationary materials, such as bonded silica gel with −CN, −NH2, or diol groups, in combination with different elution solvents for lipids separation [78]. For example, aminopropyl cartridges are used for sequential extraction of neutral and acidic phospholipids. With the application of solvents with increasing polarity, efficient isolation of phosphatidylcholine, non-esterified fatty acids, cholesterol esters and triacylglycerols can be achieved [79]. These cartridges can also be used for fractionation of the subclasses lipids, including ceramides, glycosphingolipids, sphingomyelins and phosphorylated sphingoid bases [80]. Compared to liquid-liquid extraction, solid-phase extraction show an improved selectivity and recovery for phospholipids, including phosphosphingolipids and glycerophospholipids [81]. Due to its simplicity in operation, reduced cost in solvents and easy automation, the popularity of solid-phase extraction is growing, especially in targeted lipidomics, whereas liquidliquid extraction is more suitable for non-targeted lipid profiling.
Lipids are widely distributed both inside and outside the cells. When whole-cell extracts are used, all information on the spatial distribution is lost [27]. Thus, for identification of lipids in organelles or membranes, cellular fractionation is critical [82]. In this regard, protein markers will allow for the verification of subcellular organelles, thus aiding in the preparation of lipid fractions [83]. Furthermore, the amount and composition of lipids can be highly variable between different types of tissues/cells and at different time during the day [84,85]. For example, circulating fatty acids display remarkable circadian patterns [86]. Thus, the temporal differences of lipid concentrations require time course experiments and the selective enrichment of lipid species when they are of low abundance. In this regard, metabolic labelling with chemical isotopes of lipid precursors followed by extraction and subsequent analysis is a powerful way to study kinetics of incorporation and turnover of lipids [87]. For example, fatty acid synthesis can be determined by the incorporation of stable isotope (D or 13C) -labeled precursors into lipids, such as palmitate, phosphatidylcholine and triglycerides, which are then analyzed by mass isotopomer distribution analysis [88-92]. During sample processing, chemicals can modify lipids. Oxidation of polyunsaturated fatty acyls in glycerophospholipids by reactive oxygen species leads to the formation of hydroxyls, hemiacetals and furans. Oxidized lipids are short-lived, thus complicating analytical capture [93]. In addition, the proportion of fatty acyls differs dramatically between organs. The brain is very rich in polyunsaturated fatty acyls, such as arachidonic acid (C20:4) and docosahexaenoic acid (C22:4), whereas the liver contains primarily saturated and monounsaturated fatty acyls. Depending on the organ and/or cell types, oxidation produces different lipid reaction products, which influence the discovery process of lipid biomarkers [94]. A major challenge would be to process the lipid samples without changing their structures and local localizations.
Separation and Analysis of Lipids
Fatty acid analysis demands high chromatographic resolution to separate those of various chain lengths and to accurately identify geometric and positional isomers. The most commonly adopted methodology for fatty acid separation is gas chromatography (GC) of fatty acid methyl esters (FAME) [95]. GC is a reliable tool for the quantitative analysis of complex mixtures of fatty acids. The method dates back to the discovery in 1950s that short-chain fatty acids can be separated by vapor-phase chromatography [96]. Later on, the protocol was improved by converting the long-chain fatty acids to methy esters for analysis by GC [97]. GC coupled with mass spectrometry (GC/ MS) has now become a routine procedure with broad application to biochemical, biomedical, forensic, agricultural, environmental and industrial research. GC with polar columns is suitable for the separation of complex fatty acid mixtures. For GC analysis, the polar carbonyl groups of fatty acids must first be converted to produce more volatile non-polar fatty acid methyl esters (FAMEs) [98]. Analytical laboratories frequently apply flame ionization detection (FID) for quantification of FAMEs. However, FID does not provide information on molecular mass or other structural characteristics for absolute discrimination. MS coupled with electron impact ionization (EI) helps to solve these problems. Moreover, the sensitivity and selectivity of MS methods are particularly advantageous for FAME determination in complex biological samples. Thus, the capacity to combine spectrometric examination and quantitative determination advances of GC-MS is a powerful alternative to GC-FID for FAME analysis [99].
For GC analysis of lipids, derivatization is usually done to substitute the polar groups, such as –COOH, -OH, -NH, and –SH, in order to improve their volatility and thermal stability. A wide range of derivatization reagents are available for reactions including alkylation, acylation and silylation [100-103]. Alkylation reagents modify compounds containing acidic hydrogens, such as carboxylic acids and phenols, which are converted into either esters or ethers [102]. In acylation reactions, compounds containing a labile hydrogen are transformed into esters, thioesters and amides. Acylation is commonly used to add fluorinated groups to molecules for analysis by electron capture detectors (ECD). However, due to the presence of residual acids, a purification step is required before injecting the products into GC [103]. Silylation produces more volatile and thermally stable derivatives that can be injected directly into GC. The reaction replaces the labile hydrogen of acids, alcohols, thiols, amines, amides or enolizable ketones and aldehydes with a trimethylsilyl group through nucleophilic attack (SN2) [100]. When the carboxyl group is derivatized with reagents containing a nitrogen atom, the molecule is ionized in the mass spectrometer with the nitrogen atom carrying the charge. Radical-induced cleavage can occur along the alkyl chain and gives a series of relatively abundant ions. Diagnostic ions tend to be generated at the sites of double bonds or other functional groups [20,104].
For quantification, a mixture of internal standards is added to the sample prior to extraction. Each lipid species is quantitated from respective standard curves, using the ratio between analyte peak area and corresponding internal peak area, which converts the instrumental signal into absolute amounts [95]. The absolute quantification depends on the signal strength of a single internal standard, usually heptadecanoic acid. Mammals cannot make heptadecanoic acid, but can take up fatty acids with odd-numbered chains through the diet, which interfere with the quantification. In GC/MS, fatty acids are reliably quantitated by the stable isotope dilution method, in which each analyte is compared to a deuterated analog with similar chemical and structural properties [95]. Moreover, by combining with selected ion monitoring (SIM), a high sensitivity and specificity can be achieved by GC/MS analysis. Intact molecular ions and/or large fragments are essential for identification and quantification. EI generates positive ions and extensive fragmentation. Thus, a limitation of GC/MS with EI is the fragment-rich spectra with low intensities of molecular ions. Moreover, the mass spectra rarely contain ions indicative of structural features, such as the positions of double bonds [105,106]. As an alternative, negative chemical ionization (NCI) of pentafluorobenzyl derivatives is applied [95]. However, NCI is a less robust ionization method compared with EI and needs to be used in combination with certain types of ester derivatives [92]. The NCI GC–MS has been demonstrated as a sensitive technique to determine cytochrome P450 metabolites of arachidonic acid [107].
The analysis of geometric isomers is challenging due to their subtle differences in chemical and physical characteristics. Separation of geometrical isomers in complex samples is most effectively achieved by GC using long capillary columns coated with highly polar cyanopolysiloxane stationary phases [108]. These columns enable the separation of geometric and positional isomers that are not fully resolved on columns with less polar stationary phases. However, polar columns required for separation of geometric isomers can cause co-elution of some saturated and unsaturated fatty acids, thus confounding component identification. Some of these concerns can be addressed by two-dimensional GC but this requires a complex technical arrangement [95]. Alternatively, a pre-purification step using silver-ion solid phase extraction with silver-ion impregnated silica can be employed to fractionate cis and trans isomers of fatty acids prior to capillary GC analysis [109]. Nevertheless, the temperature limit of polar columns does not permit the analysis of FAMEs with more than 30 carbon atoms.
The two soft-ionization techniques including electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) have been introduced in late 1980s and revolutionized MS field [110,111]. In contrast to EI and CI, ESI does not require the derivatization of fatty acids and generates intact molecular ions. It is particularly useful for the analysis of extremely long chain fatty acids (VLCFA) that contain up to 36 carbon atoms. The advantages of shotgun lipidomics, i.e., direct-infusion using ESI/MS for lipid analysis, are high speed and ease of automation. Using ESI, the suppression of ionization by matrix components has to be considered, particularly for direct MS approaches. Therefore, the application of internal standards is mandatory for quantification and relative comparison [89]. The addition of two internal standards for each lipid class, e.g., a low mass and a high mass species, allows the calculation of the ratios against both. By coupling ESI to liquid chromatography (LC), the number of lipid classes that can be analyzed has greatly increased. LC is applied to separate different molecular species based on the polar head groups and the different fatty-acyl chains using normal-, polar- or reversed-phase columns, or in combination. Polar stationary phases are favorable as they allow the co-elution of the lipid species of the same class and internal standards [112]. In contrast, reversed phase chromatography usually separates lipid species by chain length, thus, can be affected by matrix effects [113]. Both normal phase and hydrophilic interaction chromatography are able to differentiate bond types (ester/ether), fatty acid combinations and isobaric compounds [114]. LC increases the fidelity of lipid identification by the separation of isobaric or quasi-isobaric species [115,116]. However, the resolution of isometric species is still limited and the problems of ion 7suppression by certain lipids are common. MS/MS instruments including two mass analyzers separated by a collision cell either in space or in time allow the generation of fragment ions of lipids for identification of previously uncharacterized lipids and discrimination between lipids with similar masses and chemical structures. Fragmentation of an ion of interest allows identification of the lipids with high fidelity and a detailed understanding of “bonding” between the different building blocks (such as fatty acids, sphingoid bases, and head groups). Species with low abundance are often analyzed by interfacing MS/MS with liquid chromatography (LC–MS/MS). Beam-based MS/MS instruments include triple quadrupoles MS, ion trap MS, quadrupole-time-offlight MS and time-of-flight MS. These advances in MS form a basis for ‘‘shotgun’’ lipidomics in which precursor lipids are determined based on characteristic fragment ions [116]. Moreover, MS/MS approaches are now available for targeted analysis of different classes of lipids in complex mixtures [117,118]. For example, in untargeted lipidomics, the crude lipid extracts produce complex mass spectra with many different ionized lipids and isobaric species. Head group labeling with D9-choline and D4-ethanolamine combined with direct infusion ESI-MS/MS allows rapid analysis of phosphatidylcholine and phosphatidylethanolamine species in crude lipid extracts from liver cells [25]. It should however be noted that the detection of lipid species of very low abundance remains a major challenge. More sophisticated applications are to be developed for distinguishing isomeric (same chemical formula but different structures) and isobaric (ions with same mass) species in comprehensive profiling studies of complex samples. Ozone-induced or radical-directed dissociation are applied as novel approaches to identify the positions of double bonds [119,120]. Negative ESI-MS/MS and/or nuclear magnetic resonance are useful for studying complex glycolipids [121].
In summary, due to the highly diversified features of lipids, a combination of multiple purification strategies (e.g. total, organellar, and subclass lipidomes) and profiling methods (e.g. GC/MS, LC/ MS and others) are required for a comprehensive lipidomic analysis. Because of the inclusion of lipid class-specific internal standards, lipidomics can provide absolute quantification as well as detailed structural information of a large number of different lipid classes simultaneously.
Lipoproteomic studies
Understanding lipidome often requires the investigation of molecular interactions between lipids and proteins [122,123]. For fully appreciate the biological significance and the potential pathophysiological implications of lipids, the type, degree and strength of lipid-protein molecular interactions should be carefully considered. However, despite lipids being one of the most abundant classes of cellular metabolites, our knowledge of lipid-protein interactions is rather limited, as it is not a trivial task for searching the lipid interactomes. The advancement of mass spectrometry-based Lipoproteomic platform has facilitated the development of methods for lipid-protein interactions.
Liposome sedimentation is most frequently used for qualitatively measuring interactions between lipids and proteins in the range of 1–20 μM. Combined with proteomics analysis using Nano-LC-MS/MS, novel and unexpected proteins can be identified for those bound to acidic phospholipids [124,125], embedded in the lipid nanodiscs [126,127], carried along the secretory and the endocytic pathways [128], and associated with specific lipid targets [129]. Photoreactive lipids have been successfully used in studying lipid-protein interactions. Upon UV activation, the photoreactivable groups, such as benzophenones, 3-trifluorophenyl diazirines, aryl azides, and alkyl diazirines, crosslink with the nearest protein neighbors to form covalent bonds. The labeled proteins are subsequently isolated for identification by mass spectrometry. Using this strategy, the dynamic interactions between proteins and lipids, including cholesterol, sphingolipids, phosphatidylcholine, and phosphatidylinositol have been reported [122,130-132]. The combination of clickable, photoreactive lipid probes with proteomics approach provides a powerful tool to identify unique lipid-protein interactions [133]. For example, S-palmitoylation is a post-translational modification required for the trafficking, compartmentalization, and membrane-tethering of many proteins. The compound 17-octadecynoic acid can serve as a click chemistry probe for in situ labeling, identification, and verification of palmitoylated proteins [51,134].
In principle, the above mentioned approach using clickable photoreactive probes is applicable to lipid-protein interactions in membrane. However, an important requirement would be that the photoreactivable lipid analogues are efficiently incorporated into the membrane. In this case, major challenges include the detection of membrane proteins with low expression levels and in live cells [135]. When designing an experiment using liposomes to study interactions between lipids and membrane proteins, many factors need to be taken into account, including lipid composition and molar ratios, membrane curvature, pH and salt concentrations of the buffer etc. It is important that the lipid composition and other parameters in the assay resemble as accurately as possible the relevant membrane organelle [123]. For example, if the protein of interest is at the cytoplasmic leaflet of the Golgi complex, it is important to include phosphatidylinositol 4-phosphate in the vesicles, whereas if the protein of interest localizes to the inner leaflet of the plasma membrane, phosphatidylinositol 4,5-biphosphate at the relevant density should be added into the model membranes [44].
When using sensitive mass spectrometric methods for identification and quantification of both proteins and lipids, the extraction methods should be modified to minimize the solubilization of lipids surrounding the protein [136]. For example, compared to the classical lipoprotein isolation techniques by ultracentrifugation, which is too tedious and may alter the composition of lipoproteins, fast performance liquid chromatography (FPLC) offers rapid and reproducible separation of lipoproteins by size [40]. This technique has been proven to be sensitive, reproducible and reliable for obtaining detailed cholesterol, glycerophospholipid, and sphingolipid composition of the separated lipoprotein fractions from a small amount of serum based on ESIMS/ MS analysis. The method can also be used to identify alterations of lipoprotein lipid species in disease conditions. Similarly, a protocol has been developed for identification of lipid-protein interactions in in vivo assembled complexes. Intact protein-lipid complexes containing a protein of interest fused to the tandem-affinity purification (TAP) tag are captured from cell lysates in the absence of detergent. After removing the contaminated lipids by analytical size-exclusion chromatography, lipidomics techniques such as GC- or LC-MS can then be applied to measure the co-eluted lipids for identifying the interactions with the TAP fusions [137].
Data analysis and interpretation
Data of proteomics is transformed by searching algorithms that enable the assignment of protein sequences by comparing experimental and theoretical MS fragmentation patterns of proteolytic peptides. In the case of lipids, the bioinformatics are different and the needs remain largely unmet. The readouts of untargeted lipidomics including the retention times during LC or GC separation, the mass-to-charge (m/z) ratio, the information on molecular and fragment ions. For targeted lipidomics, the data is delivered as a matrix of lipid identities (including precursors to fragment ions) and their intensities. Typical procedure of analysis includes data processing (peak integration, identification and normalization), statistics (univariate or multivariate), and integration of functional pathways (e.g., the Kyoto Encyclopedia of Genes and Genomes, KEGG). In addition to LIPID MAPS, other lipid databases and software packages have been developed, such as LipidView, MZmine 2, LipidXplorer, LipID and LipidBank [138-144]. The composite spectrum and the molecular formula of lipid entities have been launched in the Human Metabolome Database (HMDB). Thus, HMDB is used to confirm and extend lipid identifications by matching the m/z values to reference masses and taking into account the isomeric characteristics. In untargeted lipidomics study, other major metabolite databases including Madison Metabolomics Consortium Database (MMCD), Metlin and MetaboSearch are also useful [145]. Integration of the search results from several databases leads to a more comprehensive coverage. In addition, a series of software tools and databases (KGML-ED, VANTED, MZmine, and LipidDB) can be used for the processing of lipidomics data and biochemical pathway reconstruction [146]. However, manual combination of the massive search results is generally difficult and there lacks software tools that enable simultaneous search against the multiple databases and the integration of the results. Although many software packages are available for the data analysis of unlabeled lipid species from shotgun and LC–MS based approaches, the bioinformatic processing of complex data sets that are generated by stable isotope labeling is lacking. In addition, computational models to follow the dynamics of lipid species metabolism and to integrate data sets from different approaches will be very useful for generating a comprehensive picture and revealing the complexity of various lipid species. Considering the dynamic distribution of organellar lipidome, whole cell lipidomes should be de-convoluted in order to obtain information about the subcellular organization. Lipidomic changes at the cellular level can indicate changes in subcellular organelles [82].
Large-scale studies for lipid-protein interactions have been used for tackling different biological questions, cataloguing or identifying potentially novel interactions. To validate the findings, statistical analysis based on reference datasets allows the assessment of data accuracy, sensitivity, and bias of the methodology [147]. The assumption that lipids and proteins interacting with each other are from the same pathway, or in different pathways but of similar functions, allows one to provide further confirmation based on known proteomic or lipidomic networks. Computational analyses of novel interactions include sequence comparison, and structural modeling, such as in silico prediction of lipid-binding domains [148]. These approaches are either used to create training sets for machine learning (classification) or abinitio simulations [149]. Based on the structural data (in PDB, www.rcsb.org/pdb), the amino acid residue involved in an interaction could be determined [150-158]. Generally, all approaches, including the statistical analysis of high-throughput experiment and the learning and dynamic simulation of molecular interactions, are in their early stages and thus limited by the quality and scale of current lipid-protein interaction data. With the development of analytical methods and accumulation of datasets, more computational approaches could be adopted by utilizing network (i.e. pathway analysis and functional annotation) and structural (i.e. conservation analysis and building domain databases) tools. Finally, integrating the data of lipoproteome with other data, i.e. transcriptome and metabolome, would provide a more complete depiction of cellular signaling and metabolic processes. Concurrently, the resurgence in the computational approaches to extract meaningful information from the multidimensional “interactomics” data will ultimately provide a better understanding of the regulatory systems underlying lipid functions and interactions in the context of cellular and organismal physiology.
Abnormal lipid metabolism and function have been closely associated with different pathologies such as obesity, cancer, atherosclerosis, diabetes, kidney failure, arterial hypertension, neurodegenerative and neurological disorders as well as multiple sclerosis. There is an expanding number of drugs that target lipid metabolic and signaling pathways, including the well-known cholesterol-lowering agents (statins) and cyclooxygenase inhibitors. Importantly, it is usually not just the change of one particular lipid molecule, large parts of the lipidome are remodeled in a coordinated manner. Therefore, by combining different approaches for quantitative and structural elucidation, lipidomics delivers a multi-parametric read-out for disease biomarker discovery, drug development and safety assessment, as well as pathophysiological elucidations. However, one of the major challenges in lipidomics is to obtain as comprehensive information about the lipidome as possible, especially on very low abundant lipids. In order to achieve more breakthroughs in lipid research, continued developments of new analytical tools are necessary to separate isobaric and even isomeric species and improve data processing, data mining, identification and interpretation of biochemical pathways.
This work was supported in part by grants from Seeding Funds for Basic Research of the University of Hong Kong and Research Grant Council grants (HKU779712M and HKU780613M), HKSAR.