Mycobacterial Diseases
Open Access
ISSN: 2161-1068
ISSN: 2161-1068
Short Communication - (2025)Volume 15, Issue 1
Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) has been used for the identification of Mycobacterium spp. due to its speed, reliability, cost-effectiveness and high efficiency. However, the cell wall of mycobacteria is rich in lipids which makes it difficult to obtain proteins for analysis by MALDI-TOF MS. In this study, two protein extraction protocols were compared: The MycoEx, recommended by the MALDI-TOF instrument manufacturer Bruker Daltonics and the MycoLyser protocol described here in, which used the MagNA Lyser instrument for cellular rupture. Protein extraction with the two protocols was performed using the virulent strain M. tuberculosis H37Rv (NCBI NC_000962) and the results compared. The MycoLyser protocol allowed enhanced Biotyper identification of M. tuberculosis since the scores obtained with this protocol were mostly ˃1,800. The identification accuracy was 38.8% (A), 44.5% (B), 16.7% (C) for MycoLyser and 0% (A), 16.7% (B), 83.3% (C) for MycoEx. In view of these results, it is evident that the new MycoLyser protocol for mycobacterial protein extraction allows improvement of the MALDI-TOF MS method efficacy for M. tuberculosis identification.
Matrix assisted laser desorption ionization; Time of flight; Mycobacterium spp; Cellular rupture; MycoLyser
Mass spectrometry using Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF MS) has enabled the identification of a large variety of bacterial species, including environmental and pathogenic mycobacteria, with reduced time and cost [1-3].
However, the use of MALDI-TOF for identification of mycobacteria is still less satisfactory than for other bacteria, because of the requirements of sample preparation, as the cell wall of the mycobacteria is rich in lipids and contains peptidoglycans esterified with mycolic acids, which makes it difficult to obtain proteins for analysis by mass spectrometry. In this way, this study evaluated a new protein extraction protocol to identify Mycobacterium tuberculosis [4, 5].
Using the virulent M. tuberculosis strain H37Rv (NCBI NC_000962), two protein extraction protocols were compared: The MycoEx protocol and the MycoLyser protocol developed in this study. The MycoLyser protocol used the MagNA Lyser instrument (Roche Molecular Systems), an homogenizer that disrupts and simultaneously homogenizes cells by the ultrarapid shaking of 2 ml screw cap tubes containing the cell suspension and beads, which applicability has already been demonstrated previously [6]. Briefly, the equivalent of two bacterial loops was collected in 200 μl sterile ultrapure water type I and vortex homogenized. For inactivation, the bacterial suspension was incubated at 95°C for 45 minutes and cooled at room temperature before addition of 700 μl of absolute ethanol. For cell disruption, 0.5 mm zirconium/silica beads (BioSpec Products) were added, followed by homogenization in MagNA Lyser apparatus for three cycles of 30 seconds at 5,000 rpm [7].
The supernatant was transferred to another tube and after centrifugation for five minutes at 14,000 × g, the supernatant was discarded and the pellet incubated for three minutes at room temperature to dry the remaining ethanol. Then, 10 μl of formic acid were added followed by an equal volume of acetonitrile for protein extraction, which were retrieved in supernatant after 15 seconds vortexing and two minutes centrifugation at 14,000 × g. Subsequently, 1 μl of the supernatant was applied to the well of the MALDI-TOF plate (MTP 384 ground steel, Bruker Daltonics) and allowed to air dry at room temperature [8]. Upon addition of 1 μl of α-cyano-4- hydroxy-cinnamic acid (5 mg/ml) in a solution containing 50% acetonitrile and 2.5% trifluoroacetic acid (v/v), crystallization of the matrix-analyte mixture was accomplished after air dry at room temperature. 18 replicates of this crystallized mixture were analyzed in an Autoflex III Smartbeam mass spectrometer (Bruker Daltonics) and obtained spectra were processed using the MALDI Biotyper 3.1 program (Bruker Daltonics) with the standard configurations. For identification, the protein profiles detected were compared to the reference library IVD (Bruker Daltonics) containing BDAL (7,311 spectra) and Mycobacterium (912 spectra) libraries. The Mann-Whitney statistical test was used to compare the protocols [9].
When comparing the MycoEx and MycoLyser protocols for protein extraction using the H37Rv reference strain of M. tuberculosis as standard, it was observed that the highest identification scores with the Biotyper software were obtained when using the MycoLyser protocol. The medians of the 18 spectra obtained experimentally were 1.656 for MycoEx and 1.862 for MycoLyser, being significantly higher for MycoLyser protocol. In addition, the MycoLyser scores were mostly higher than the 1,800 limit proposed by Bruker Daltonics as the minimum value for mycobacteria identification with high confidence (Figure 1) [10].
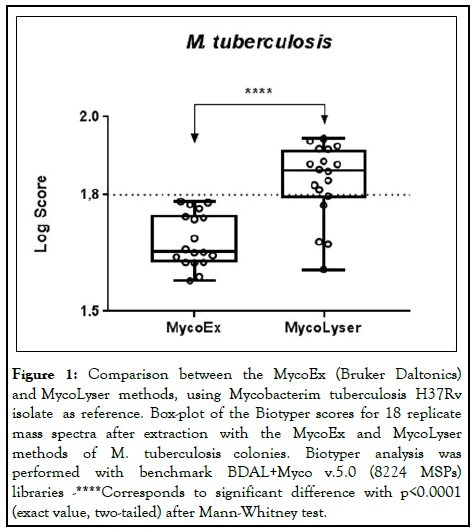
Figure 1: Comparison between the MycoEx (Bruker Daltonics) and MycoLyser methods, using Mycobacterim tuberculosis H37Rv isolate as reference. Box-plot of the Biotyper scores for 18 replicate mass spectra after extraction with the MycoEx and MycoLyser methods of M. tuberculosis colonies. Biotyper analysis was performed with benchmark BDAL+Myco v.5.0 (8224 MSPs) libraries -****Corresponds to significant difference with p<0.0001 (exact value, two-tailed) after Mann-Whitney test.
It was also found that the MycoLyser protocol provided better results regarding the accuracy of the identification, since the mass spectra obtained with this protocol were more frequently correctly identified with M. tuberculosis. The Main Spectra Profiles (MSPs) generated with the mean peak frequency of the 18 replicates obtained with MycoEx and MycoLyser protocols were compared with the reference library (BDAL+Myco v.5.0), containing 8223 different bacterial MSPs, 912 of which being mycobacteria reference spectra, including species of the Mycobacterium Tuberculosis Complex (MTC) [11].
The Main Spectra Profiles (MSPs) generated with the mean peak frequency of the 18 replicates obtained with MycoEx and MycoLyser protocols were compared with the reference library (BDAL+Myco v.5.0), containing 8223 different bacterial MSPs, 912 of which being mycobacteria reference spectra, including species of the Mycobacterium Tuberculosis Complex (MTC). In this analysis, the highest score of identification obtained for MycoEx protocol MSP was Mycobacterium bovis (Bovinus An_1 PGM) wrong match, whereas for MycoLyser protocol MSP, the highest score was Mycobacterium tuberculosis_complex (reference M. tuberculosis 03L LDW b) correct match (Figure 2) [12,13].
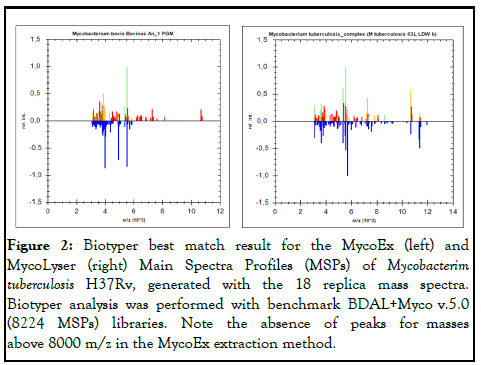
Figure 2: Biotyper best match result for the MycoEx (left) and MycoLyser (right) Main Spectra Profiles (MSPs) of Mycobacterim tuberculosis H37Rv, generated with the 18 replica mass spectra. Biotyper analysis was performed with benchmark BDAL+Myco v.5.0 (8224 MSPs) libraries. Note the absence of peaks for masses above 8000 m/z in the MycoEx extraction method.
The three highest scores obtained with the 18 MycoEx and MycoLyser spectra were also evaluated for identification reliability. Of the 18 MycoEx spectra, only 8 (44.44%) presented the highest identification score being M. tuberculosis, whereas with MycoLyser this was observed for 14 spectra (77.77%). We also verified that 11 of the 18 MycoLyser spectra (61.11%) had the first three results (first, second and third highest scores) listed as M. tuberculosis, whereas for MycoEx this could not be observed [14].
According to Bruker Daltonics, the reliability (high, medium and low) of identification can be evaluated by the consistency of the three highest scores for identification for the same genus and/or species. Those three categories were adopted here, as following: A=Species consistency, in which the reliability of species identification is high; in this case the three highest scores must present the same identification result for gender and species; B=Gender consistency, when only gender identification is highly reliable; when the three highest scores show the same gender identification result; C=Absence of consistency for species or genus, in which neither gender nor species show reliable identification since criteria A and B described above are not met. The consistency of the MycoLyser method resulted in 38.8% (category A), 44.5% (category B), 16.7% (category C) whereas the MycoEx method resulted in 0% (category A), 16.7% (category B) and 83.3% (category C) [15].
Interestingly, the extraction of proteins with the MycoLyser protocol also allowed greater detection of peptides and proteins with mass values above 8,000 Daltons (Figure 2), which may help to explain the higher confidence and reliability of MycoLyser protocol for M. tuberculosis identification by MALDITOF MS [16].
Although MALDI-TOF MycoEx protocol accurately classifies MTC isolates at the genus level, the differentiation of M. tuberculosis and other species within the MTC has not been described yet [17].
While it is not the objective of this study, our results suggest that MTC mycobacteria identification at species level can be accomplished by MALDI-TOF MS and the MycoLyser protocol usage may be an important tool within this context. In view of these results, it is evident that the new MycoLyser protocol for mycobacterial protein extraction allows improvement of the MALDI-TOF MS method efficacy for M. tuberculosis identification.
To the researchers Carlos Bloch Jr and Daniel Sifuentes, for technical support at the laboratory of mass spectrometry of embrapa genetic resources and biotechnology. This study was supported by the following grants: Embrapa 02.13.10.008.00.00 and 03.13.10.008.00.00, Fundect 085/2015, 59/300, 121/2015 and 071/2017 and CNPq 407826/2018-1 and 443235/2014-7.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Bacanelli GM, Araujo FR, Verbisck NV (2025) MALDI-TOF Protocol for M. tuberculosis Identification Improved MALDI-TOFMS Identification of Mycobacterium tuberculosis by Use of a Cell Disruption Protocol. Mycobact Dis. 13:359.
Received: 25-Sep-2019, Manuscript No. MDTL-23-2336; Editor assigned: 30-Sep-2019, Pre QC No. MDTL-23-2336 (PQ); Reviewed: 14-Oct-2019, QC No. MDTL-23-2336; Revised: 01-Nov-2023, Manuscript No. MDTL-23-2336 (R); Published: 29-Jan-2025 , DOI: 10.35248/2161-1068.23.13.359
Copyright: © 2025 Bacanelli GM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.