
Journal of Clinical Trials
Open Access
ISSN: 2167-0870

ISSN: 2167-0870
Research Article - (2022)
The ongoing SARS-CoV-2 pandemic has disrupted health systems worldwide. SARS-CoV-2 is constantly evolving and multiple newer Variants of Concern (VOC) such as Delta and Omicron have emerged. Therefore, there is a need for drugs which can be achieved by targeting the proteins that are conservative and less susceptible to mutations and can initialize T cells activation. The viral polymerases such as PLpro (nsp3), 3CLpro (nsp5), and RdRP (nsp12), proteins critical for viral replication, are highly conserved in RNA viruses. Targeting this unique subset of viral replication proteins may prove effective against a wide range of VOC. Notably, when T cells encounter an antigen that their receptors recognize, they undergo self-replication and produce more immune cells. Hence, the immune characterization and identification of the T-cell immune epitopes for key viral replication protein RdRp (RNA-dependent RNA polymerase) is very important. Previously, we showed the efficacy of Curvic®, a combination of phytoactives in the health management of COVID patients. Various preclinical and clinical studies conducted with Curvic® have shown an increase in T cell counts and overall improved immune response in COVID patients, however, the mechanism was unclear. Herein, we provide data to show that Curvic® inhibits the conservative replication protein RdRp (nsp12), and thereby prevents viral replication and reassembly. Our studies show that Curvic® increases T-cells that have immune epitopes for RdRp (nsp12). Further, we believe that increased T cell count in blood by Curvic® treatment might be the result of T cells selfreplication to produce more immune cells upon encountering and recognizing fragments viral protein RdRp. While, some of these T cells instantly target and destroy infected cells, others may circulate within the body to combat the same infection in the event of its re-emergence and suggest Curvic® to be a promising intervention against SARS-CoV-2, MARS and other Coronavirus infections. The same mechanism can also be attributed to the other viral diseases.
SARS-COVID; T cells; RNA viruses; Spike glycoprotein
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-Cov-2) is a positive Sensed Single Stranded RNA (+ssRNA) [1]. The genomic RNA of SARS-CoV-2 includes ten ORFs (Open Reading Frames), amongst which two major ORFs: OFR1a and ORF1b encompasses two-third of the viral genome and are translated into proteins: pp1a and pp1b, crucial for encoding polypeptides responsible for the replicase-transcriptase complex of the virus [2,3]. The remaining one-third of the viral genome consists of overlapping ORFs, largely involved in the encoding of main structural proteins, including: Membrane protein (M), Envelope protein (E), Spike glycoprotein (S), Nucleocapsid protein (N) and other accessory proteins [4,5].
The virus enters by binding to the host cell ACE2 receptor (Angiotensin-Converting Enzyme 2) with the help of Spike protein (S) [6,7]. After the interaction between the S protein and ACE2 receptor, the S protein undergoes a critical proteolytic cleavage and subsequent fusion with the host membrane, thereby initiating the viral infection [8]. Upon entering the cell, virus releases the ssRNA into the cytoplasm. The viral RNA replicates inside the host cell. The replication and transcription of the viral genome are regulated by the assembly of Non- Structural Proteins (NSP) that assembles to form RNA-synthesis complex and initiate replication. NSP12 RNA-dependent RNA polymerase (RdRp) is the key catalytic subunit which along with co-factors nsp7 and nsp8 carries out the nucleotide polymerization and replication of SARS-CoV genome. After quick replications, the virus invades the host cell system and enters the blood stream [9].
The antigens present on the surface of the virus initiate the bodies humoral and cellular immune response mediated by virus-specific T and B cells. According to the reports, the SARSCoV- 2 infection decreases the count of CD4+ and CD8+ T cells in the peripheral blood. These cells are further characterized by hyper-activated appearance evidenced by the excess proportion of double-positive fractions of HLA-DR (CD4 3.47%) and CD38 (CD8 39.4%) [10]. The virus is also responsible for inducing T-cell exhaustion showing high levels of PD-1 and Tim-3 exhaustion markers [11]. For the efficacy of the vaccines, recognition of antigen is crucial for eliciting an immune response and subsequent killing.
Spike proteins have been the principal target of medicines and vaccines (mRNA and adenovirus-based) licensed for use for the patient's treatment following infection by SARS-CoV-2 [12]. However, multiple newer Variants of Concern (VOC) such as Delta and Omicron, have emerged, whose spike proteins carry numerous mutations enabling their immune evasion and survival in the host system without getting targeted. These spikeborne mutations make it difficult for antibodies and immune cells (triggered by vaccination) to identify the variants, thus reducing the efficacy of these preventive measures [13]. Therefore, there is a need for treatments/vaccines that are more extensive and with robust immune responses. This can be achieved by targeting the proteins that are less susceptible to mutations and can initialize T cells activation.
Notably, unlike the spike proteins that constantly evolve by mutations, Nonstructural Proteins (NSP) of SARS CoV-2 viruses such as viral polymerases, PLpro (nsp3), 3CLpro (nsp5), and RdRP (nsp12) play an important role in viral replication and are conserved proteins [14]. The computational analysis reveals the active site of the RdRp (RNA-dependent RNA polymerase, nsp12) protein to be a highly conserved and accessible region [15,16]. Hence targeting 3CLpro (npro) and RdRp protein for inhibition of viral replication can be an effective therapeutic approach to disrupt the viral life cycle. Studies by Sette et al., have shown that people who have been infected with SARSCoV- 2 typically generate T cells that target at least 15-20 different fragments of coronavirus viral replication proteins [17]. Cell-mediated T-cell immune epitopes have been identified for RdRp protein using immunoinformatic techniques, which is proved to be a rapid and efficient method to explore the RdRp (nsp12) target as an effective candidate [9,18].
Adaptive immune responses are important for the control and clearance of viral infections and comprise activities of B cells, CD4+ T cells, and CD8+ T cells [18,19]. Importantly, surfaces of T-cells contain receptors that recognize antigens i.e. foreign protein fragments. When T cells interact with an antigen that is identified by their receptors, they undergo self-replication and produce more immune cells. Some of these T cells instantly target and destroy infected cells, while others circulate within the body to combat the same infection in the event of its reemergence and therefore are an integral part of the adaptive immune response [9]. A strong correlation between SARSCoV- 2-specific CD4+ T and CD8+ T cells and severity of COVID-19 disease was reported; higher the number of CD4+ and CD8+, minimal was the damage after the SARS-CoV-2 infections [20,21]. These findings suggest that by stimulating CD4+ and CD8+ T cells in the body, COVID-induced health complications can be averted.
In a recent study, two RdRp epitopes were found to activate broad coronavirus reactive T cell responses [18,19]. Cytotoxic analysis revealed that most of the TCRs recognized and killed RdRp expressing cells. Additionally, the TL9 (an epitope) reactive TCR exhibited strong cross-reactivity with SARS, MERS, 229E, and NL63 coronaviruses. These studies identify targeting of RdRp as an effective strategy to combat virus associated cytotoxicity and health complications.
Recently “Curvic®”, a formulation made from natural molecules was shown to help in developing immunity against SARS-CoV-2 (COVID-19) infections [22-24]. The main ingredients of Curvic® are Curcumin, special proprietary form of curcumin vitamin K2-7, Vitamin C, Zinc and L-Selenomethionine. The preclinical efficacy and safety studies conducted on Curvic® showed Curvic® to be safe [23]. Moreover, the immune response triggered by Curvic® comprised a remarkable increase in CD4+ counts on average by 75.75% and CD8+ counts by 64.44%. The proposed mechanism of Curvic® protection against SARS CoV2 infection and activation of the immune system have been linked to inhibition of COVID-19 Mpro protein [24]. To have further insight into the protective mechanism of Curvic® and whether it targets conservative viral proteins, we conducted computational analysis using SARS-CoV-2 target proteins and active ingredient of Curvic®. Interestingly, the new data indicates that the phytoactives from Curvic® are inhibitors of RdRp (nsp12), along with the inhibition of Mpro protein inhibition [24].
To predict the interactions between active ingredient of Curvic®, Prepinim which is a modified curcumin structure and the target SARS-CoV-2 proteins, docking analysis was performed. Docking is an established in silico technique to explore the sites between drug and targets that can serve as a potential candidate. For computational analysis, 2D structural information of the active compound of Curvic® was retrieved from The National Library of Medicine: Curcumin-PubChem CID: 969516. For docking analysis, the crystal structures of major SARS-CoV-2 target proteins were retrieved from Protein Data Bank (PDB), shown in Table 1. Using Auto dock tools along with the information regarding the active sites of the proteins from the literature, docking analysis was performed. The parameters used for studying interaction between Curvic® components and each viral protein included {Total number of conformations=20, Number of points in X-dimension=25, Number of points in Ydimension= 25, Number of points in Z-dimension=25}. Interactions were analyzed by the Autodock program and wherever Auto dock couldn't read analysis was performed by Biovia Discovery Studio 4.5. Auto dock software generated potential interaction sites and values for binding energy that predict the affinity between Curvax molecules and SARS CoV-2 target proteins, selected in this study.
| PBD Id | Target | Analyzed by | |
|---|---|---|---|
| 1 | 5X29 | E protein pentameric ion channel | Autodock |
| 2 | 6JYT | Nsp13 | Autodock |
| 3 | 6LU7 | Main protease | Autodock |
| 4 | 6VW1 | SARS-CoV-2 chimeric receptor-binding domain that binds to receptor ACE2 | Autodock |
| 5 | 6VWW | NSP15 Endoribonuclease | Autodock |
| 6 | 6WKQ | NSP16-NSP10 Heterodimer | Autodock |
| 7 | 6WZU | Papain-Like Protease | Autodock |
| 8 | 6VSB | Spike glycoproteinwith a single receptor-binding domain | Discovery studio |
| 9 | 6VXX | Spike glycoprotein (closed state) | Discovery studio |
| 10 | 6VYB | Spike ectodomain structure (open state) | Discovery studio |
| 11 | 7BV2 | RdRp (nsp12-nsp7-nsp8 complex-RNA site) | Discovery studio |
Table 1: Docking of Curvic® compounds with potential COVID-19 drug targets.
Replication of SARS-CoV-2 virus involves the coordinated function of several structural and Nonstructural Proteins (NSPs). NSP12 is the catalytic component of RNA-dependent RNA polymerase (RdRp), important in the regulation of viral replication. Notably, NSP12 function is dependent on many other proteins such as NSP7 and NSP8. The detailed comparative docking analysis of some of the key targets with active compounds of Curvic® is provided below.
Docking analysis of 5X29 (E protein pentameric ion channel) with Curvic® component
Docking between 5X29 and curcumin was performed using Auto Dock Tools as explained in the methods. Docking of 5X29 with curcumin shows no interaction with the binding energy for the first conformation observed as -5.2 Kcal/mol details shown in Table 2 and Figure 1.
| PBD Id | Target | Interactions: Bond length/Binding energy | |
|---|---|---|---|
| 1 | 5X29 | E protein pentameric ion channel | no interaction/-5.2 |
| 2 | 6JYT | Nsp13 | ILE20: HN→ O3/-5.4 |
| 3 | 6LU7 | Main protease | GLY143: HN→ O2/-6.8 |
| 4 | 6VW1 | SARS-CoV-2 chimeric receptor-binding domain that binds to receptor ACE2 | TYR505: O→ H13/-6.7 GLN409: HE22→ O4/-6.7 ILE418: HN→ O4/-6.7 |
| 5 | 6VWW | NSP15 Endoribonuclease | LYS71: HZ2→ O2/-8.7 |
| 6 | 6WKQ | NSP16-NSP10 Heterodimer | CYS6913: HN→ O3/-8.7 |
| 7 | 6WZU | Papain-Like Protease | ARG138: HH2→ O5/-6.8 |
| 8 | 6VSB | Spike glycoprotein with a single receptor-binding domain | interaction not in active site pocket/-5.0 |
| 9 | 6VXX | Spike glycoprotein (closed state) | interaction not in active site pocket/-5.4 |
| 10 | 6VYB | Spike ectodo main structure (open state) | GLN52:HE22-O3/-4.8 |
| 11 | 7BV2 | RdRp (nsp12-nsp7-nsp8 complex-RNA site) | ARG624:HH22-O1, ASN691:HD21-O5, ASN691:HD22-O6/-7.0 |
Table 2: Analysis of active compound (Curcumin) of Curvic®.
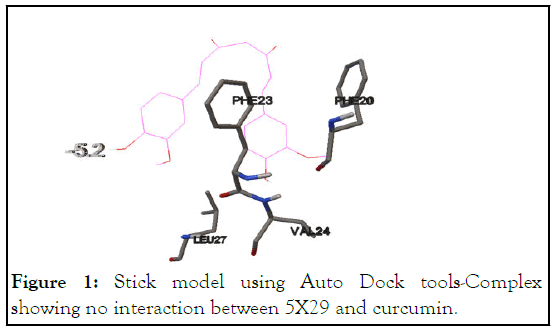
Figure 1: Stick model using Auto Dock tools-Complex showing no interaction between 5X29 and curcumin.
Docking analysis of 6LU7 (Main protease with Curvic® component
Docking analysis of 6LU7 (SARS-CoV-2 main protease) with Curcumin shows that the interaction is taking place at residue-GLY143 (the predicted active site) with the binding energy of -6.8 kcal/mol. Another interaction between 6LU7 and GLY143 occurs but with different binding energy of -5.9 kcal/mol and suggest that SARS-CoV-2 main protease is definitely one of the targets of Curvic®, shown in Table 2 and Figures 2 and 3.
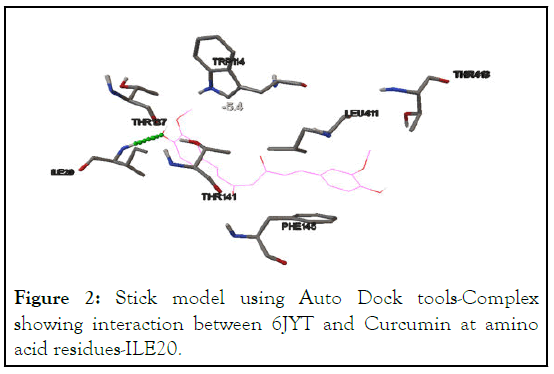
Figure 2: Stick model using Auto Dock tools-Complex showing interaction between 6JYT and Curcumin at amino acid residues-ILE20.
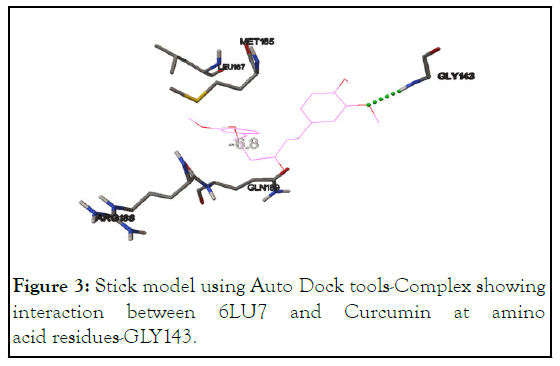
Figure 3: Stick model using Auto Dock tools-Complex showing interaction between 6LU7 and Curcumin at amino acid residues-GLY143.
Docking analysis of 6VW1 (SARS-CoV-2 chimeric protein that binds to receptor ACE2) with Curvic®
Docking analysis of 6VW1 with Curcumin using Auto Dock Tools, reveal interactions at residues-GLN409, ILE418 and TYR505 with the binding energy of -6.7 kcal/mol (at the predicted active site). Curcumin was found to interact with 6VW1 protein at multiple amino acid residues, suggesting that SARS-CoV-2 chimeric protein is a strong target for Curvic® (Table 2 and Figure 4).
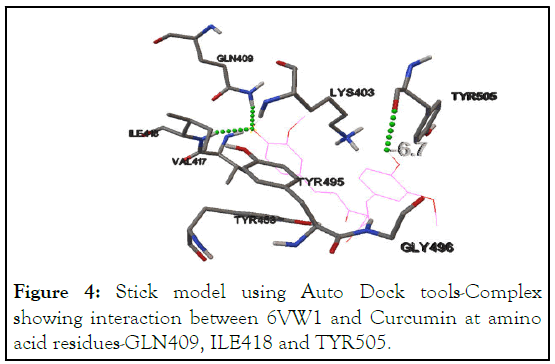
Figure 4: Stick model using Auto Dock tools-Complex showing interaction between 6VW1 and Curcumin at amino acid residues-GLN409, ILE418 and TYR505.
Docking analysis of 6VWW (NSP15 Endoribonuclease of SARS-CoV-2) with Curvic®
6VWW interacted with curcumin at the residue-LYS71 with the binding energy of -8.7 Kcal/mol. All the interactions occurred at the predicted active sites, shown in Table 2 and Figure 5, shown in Table 2 and Figure 6.
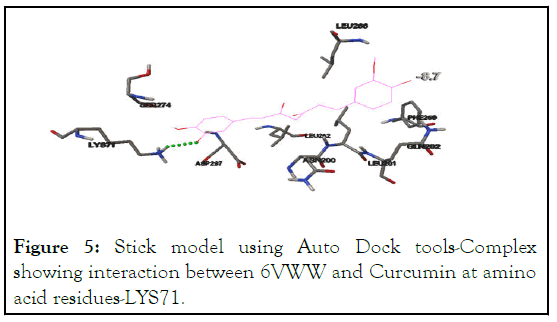
Figure 5: Stick model using Auto Dock tools-Complex showing interaction between 6VWW and Curcumin at amino acid residues-LYS71.
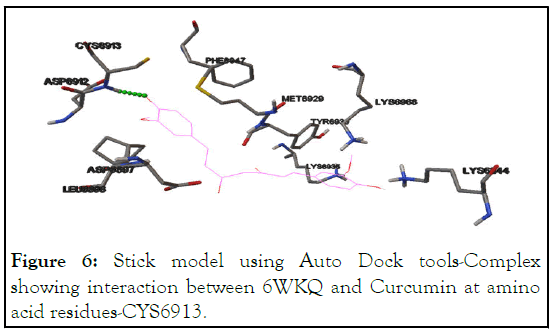
Figure 6: Stick model using Auto Dock tools-Complex showing interaction between 6WKQ and Curcumin at amino acid residues-CYS6913.
Docking analysis of 6WZU (Papain-Like Protease) with Curvic®
Docking of 6WZU with Curcumin identify residue-ARG138 to be an interaction site with the binding energy of -6.8 kcal/mol. ARG138 is the predictive active site, shown in Table 2 and Figure 7.
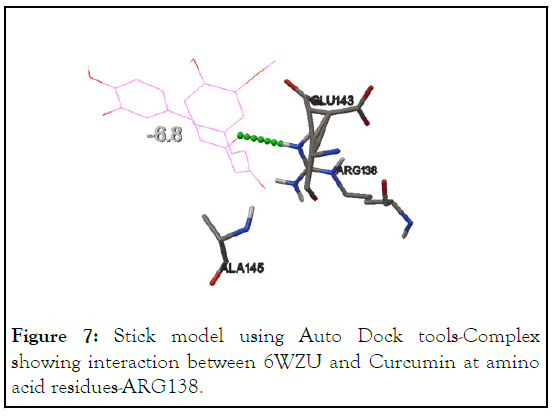
Figure 7: Stick model using Auto Dock tools-Complex showing interaction between 6WZU and Curcumin at amino acid residues-ARG138.
Docking analysis of 7BV2 (SARS-CoV-2 nsp12-nsp7- nsp8 complex with Curvic®
Docking of 7BV2 with Curcumin using Discovery studio tools revealed interaction at multiple sites including ARG624, ASN691, and ASN691 with binding energy -7.0 kcal/mol, suggesting that Curvic® has strong affinity for nonstructural protein NSP12-complex, shown in Table 2.
Overall, our in silico computational analysis identified several interactions between Curvic® compound and active sites of the selected SARS-CoV-2 targets proteins, including main protease, Papain like protease and NSPs (NSP12-NSP7-NSP8 complex, NSP 13, NSP15 and NSP 16-NSP10). Most of these Curvic® interacting proteins are involved in the replication or assembling process of the SARS-CoV-2 virus. Interestingly, the molecule of Curvic® interacted with RdRp (NSP12 complex) at multiple amino acid residues, suggesting that RpRd is a definite target for Curvic®. Curvic® was found to be effective against COVID infection in the clinical study [22]. In this randomized two-arm study, 200 COVID-19 positive patients were enrolled and were given either Curvic® or standard treatment for 10 days. Within 2 days of Curvic administration, a significant drop in temperature to afebrile level was observed, whereas in patients who received standard treatment, it took 4 days for fever to drop to afebrile levels. Moreover, the group that received Curvic® displayed improved T cell count, and better recovery. Based on the present study, we attribute the improved immune response and beneficial effects of Curvic® seen in the clinical setting to its ability to interact with viral proteins involved in the replication and assembling of the virus as described by Sette et al. [17,18]. According to them, adaptive immune system play critical role against SARS-CoV-2 and that there is a direct correlation between T cells generated by patient’s immune system and severity of the COVID disease. They further explained that generation of T cells that recognize the viral fragments of coronavirus, kill and remove the virus from the body [17]. Since our docking data shows strong affinity between Curvic® and RdRp and other proteins involved in replication of virus, we believe that Curvic® interaction with RdRp might lead to fragmentation and subsequent identification by infected patient’s adaptive immune system as explained by Sette et al. [17]. Further, recognition of the viral proteins might have triggered T-cell count and subsequent removal of the virus as observed in the patients who received Curvic® in the clinical study [25].
Based on our docking data, it can be concluded that Curvic® formulation is able to interact with viral proteins and block the replication of the virus possibly by blocking the RdRp (nsp12) and viral assembly proteins followed by recognition of fragmentation of RdRp and 3CLpro proteins process by the TCR which further induces an immune response. Our data suggest that use of Curvic® will not only minimize the level of severity of the disease but also suppress the viral spread by preventing replication. Therefore Curvic® formulations present huge immunomodulatory potential against SARS-CoV-2 infection and we believe, this response is not restricted to SARS-CoV-2 only, but may also be effective against MARS, Coronavirus and Adenovirus.
Author is thankful to M/s SSV Phytopharmaceuticals for their funding support.
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
Citation: Dound YA (2022) Mechanistic Understanding of Novel Herbomineral Formulation Curvic® in the Management of Viral Outbreaks. J Clin Trials. S20:007
Received: 23-Aug-2022, Manuscript No. JCTR-22-18977; Editor assigned: 26-Aug-2022, Pre QC No. JCTR-22-18977(PQ); Reviewed: 12-Sep-2022, QC No. JCTR-22-18977; Revised: 19-Sep-2022, Manuscript No. JCTR-22-18977(R); Published: 26-Sep-2022 , DOI: 10.35248/2167-0870.22.S19.007
Copyright: © 2022 Dound YA. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.