Research Article - (2016) Volume 2, Issue 1
Microbial Diversity of Ammonia Oxidizing Bacteria through Waste Water Genomics
*Corresponding Author: Maulin Pramod Shah, Industrial Waste Water Research Laboratory, Division of Applied and Environmental Microbiology, Enviro Technology Limited, Gujarat, India, Tel: 91-9099965504 Email:
Abstract
Autotrophic ammonia-oxidizing bacteria are a critical factor of the microbial community in industrial wastewater treatment systems. We evaluated the diversity and community composition of β-proteobacterial ammonia-oxidizing bacteria in two full-scale treatment reactors - a sand filter and a biological aerated filter - receive an identical wastewater. Polymerase chain reaction of the 16S rRNA gene fragments of ammonia-oxidizing bacteria-selective primers was merged with denaturing gradient gel electrophoresis to allow the comparative analysis of the dominant ammonia-oxidizing bacteria populations. The phylogenetic affinities of the dominant ammonia-oxidizing bacteria were verified by cloning and sequencing of polymerase chain reaction-amplified 16S rRNA gene fragments. Denaturing gradient gel electrophoresis profiles were evaluated using a probability-based similarity index. An exploitation of a probabilistic index of similarity permitted us to consider the differences and similarities observed in ammonia-oxidizing bacteria community structure in different samples were statistically significant or could be accounted for random matching of bands in denaturing gradient gel electrophoresis profiles that would propose random colonization of the reactors at different ammonia-oxidizing bacteria. All Possibly-like sequences recognized, grouped within the Nitrosomonas genus. A greater diversity of ammonia-oxidizing bacteria were detected in trickling filters than the BAF on all samples analyzed were initiate to be dominated by ammonia oxidizing bacteria most closely linked to Nitrosococcus mobilis. Numerical investigation of the denaturing gradient gel electrophoresis profiles indicated that the ammonia-oxidizing bacteria community in depth profiles from the filter beds was selected in a non-random manner.
Keywords: Environmental Genomics, Ammonia oxidizing bacteria, Nitrosococcus, Nitrosomonas
Introduction
Soil environment offers an amazing diversity of microorganisms, and the composition of soil microbial communities may be highly variable within the area. Despite considerable interest in understanding how microbial communities are structured across space [1-3], we still have a relatively limited understanding of biotic and abiotic factors that can drive the observed spatial variability of community composition. In addition, because the vast majority of soil bacteria were grown, we can use studies that compare the community in a variety of soil types to gain insight into the ecology, physiology and life history strategies, uncultivated microbial taxa. A number of recent studies have examined the distribution of bacterial communities in a variety of different soils. However, these studies are limited to high levels of soil bacterial diversity, which (currently) makes it difficult to survey the entire community high level taxonomic resolution over a large number of individual samples. This limitation is less problematic when considering the distribution of individual bacterial taxa that can be studied at relatively subtle levels of phylogenetic resolution. Ammonia oxidizing bacteria represented genera Nitrosomonas and Nitrosospira within β-subclass Proteobacteria to [4] are particularly suitable for investigating the distribution of soil microbes in space as phylogenetic relationships within the ammonia oxidation bacteria (ammonia oxidizing bacteria) are quite well described [5]. Furthermore, unlike many other bacterial taxa found in soil, it is a group represented by the relatively large number of cultured isolates. In addition, ammonia oxidizing bacteria, together with the ammonia-oxidizing archaea [6-8] to perform the step of limiting the rate of nitrification and play a key role in regulating the dynamics of soil nitrogen. For this reason, studies of ammonia oxidizing bacteria biogeography may have a direct relationship to study soil biogeochemical given that different ammonia oxidizing bacteria groups may have different physiological and ecological characteristics [9].
Despite the fact that the ammonia oxidizing bacteria have been studied for decades, their biogeochemical importance, ubiquity and which have the potential to serve as a "model taxon" biogeographical studies, we know of no previous studies that comprehensively investigated diversity and composition of ammonia oxidizing bacteria communities in many types of ecosystems. We know that distinct soil often conceal various ammonia oxidizing bacteria communities. A number of environmental factors, including the type of vegetation [10-12], the nutrient levels in the soil [13-17], soil microclimate [13,17,18], and control procedures [19-21] was found to have a significant effect on spatial variability exhibited ammonia oxidizing bacteria communities. However, because most studies have compared the ammonia-oxidizing bacteria communities across a rather limited number of samples, with one exception, the study by Avrahami and Conrad [18], it was difficult to determine what soil biotic and abiotic characteristics are associated with ammonia oxidizing bacteria community composition across a larger space. Of course we must be cautious about interpreting these studies because they are basically observing nature. No one has yet conducted robust, controlled and statistically designed experiments on the effect of environmental conditions on the distribution of ammonia-oxidizing bacteria. The purpose of this study was to investigate the diversity of ammonia oxidizing bacteria in two deferent full-scale reactors treating sewage same mixed domestic and industrial waste. Many culture techniques independent of it was used. Polymerase chain reaction was used to amplify fragments of the 16S rRNA gene primers with selective to ammonia oxidizing bacteria beta-proteobacterial [19,22]. PCR amplified fragments were analyzed by denaturing gradient electrophoresis gel and the profiles were numerically analyzed to statistically significant compared populations dominant ammonia oxidizing bacterial reactors. Moreover, selection of the cloned PCR - amplified fragments of the 16S rRNA gene derived from the reactors was sequenced to determine the phylogenetic affiliation prevailing ammonia oxidizing bacteria -like sequences.
Materials and Methods
Reactor operation
Samples were assembled from an industrial waste water treatment plant receiving a blend waste of domestic and industrial source. The plant is in the shape of a multi-step treatment process. Two reactors were examined from the treatment plant, a filter bed reactor and biological aerated filter operating in parallel. Typical loads of the wastewater treatment plant: TBOD, 3280 kg/day; COD: 14800 kg/day; NH4+ - N: 800 kg/day; TSS: 2860 kg/day; and typical values for final effluent quality of the facility is: TBOD: 8.0 mgL-1; COD: 122.8 mgL-1; NH+4 – N: 1.4 mgL-1; TSS: 16.4 mgL-1.
Sampling
Biomass samples were taken in a duplicate from the back- wash flow from each unit in the biologically activated filter system. Top middle and lower of the primary and secondary sample was taken from filter beds of manual excavation. Biomass was retained immediately on samples in 55 % ethanol and stored at -20°C before analysis.
Nucleic acid extraction and polymerase chain reaction /RTPCR amplification
Sediments were compacted by centrifugation at 12,000 rpm for 20 min and excess water removed. DNA were extracted and purified as described by Harris et al. from 0.5 g of compressed sediment after disruption of cells by bead-beating. Both the nucleic acids templates were prepared following resuspension of extracted nucleic acids in 60 μl of RNase-free sterile water, and DNA templates were again purified using 0.6 ml Viva spin concentrator columns. Nucleic acid extracts were quantified by standard agarose electrophoresis and polymerase chain reaction templates were adjusted to equal concentrations by dilution. Ammonia-oxidiser 16S rRNA gene fragments were amplified from extracted DNA by nested polymerase chain reaction using β- proteobacterial ammonia oxidiser primers (Hayes P. 2001) followed by the general eubacterial primers (Nicol et al. 2003), after a 1:50 dilution to prevent non-specific amplification. Planctomycete 16S rRNA gene fragments were amplified using the semi-specific Planctomycetales- Verrucomicrobia assay (Nicol et al. 2003) with the primers PLA40f and 1492r. 16S rRNA gene sections were also amplified from DNA generated from extracted RNA by reverse transcriptase- polymerase chain reaction by means of a variation of the method described by Griffiths et al. DNA-free RNA was acquired by treating 10 μl of extracted RNA with 3 U of RQ1 RNase-free DNase for 25 min at 40°C, to which 10 μl of RT reaction mixture was added according to the manufacturer’s instructions. We used 1 μl of digested nucleic acids as polymerase chain reaction-template to ensure complete removal of DNA. Reverse transcription was carried out at 42°C for 50 min and the enzyme was subsequently heat inactivated for 15 min at 70°C. Polymerase chain reaction-amplified fragments were resolved by standard horizontal electrophoresis on 1% (w/v) agarose gels. 16S rRNA gene fragments were also polymerase chain reaction -amplified from pure cultures and clones representative of β-proteobacterial ammonia oxidizing bacterial clusters as described by Nicol et al. and Griffith et al. Planctomyces maris AL and P. brasiliensis VL32 were used as controls for polymerase chain reaction amplifications with the PV assay. The primers and the polymerase chain reaction conditions for all primer pairs used are tabulated in table 1. An individual reagents and their concentrations were as follows: 1 × polymerase chain reaction buffer, 1 U Biotaq DNA polymerase and 1.5 mM MgCl2; each primer had a concentration of 0.2 μM and each deoxynucleoside triphosphate a concentration of 250 μM. Amplification was carried out with a total volume of 50 μl, using an omn-E thermal cycler and applying the thermal cycle conditions in table 1.
| Primer | Target group | PCR approach | Thermocycling programme |
|---|---|---|---|
| CTO189f-GC–CTO654 (465 bp) | β-subgroup AOBs | DGGE/nested amplification 1st stage | Initial denaturing time 5 min; annealing temperature 55°C; extension time 45 s; final extension time 5 min |
| PLA40f-pf1053r (1.0 kb) | Planctomycetales- Verrucomicrobia | DGGE/nested amplification 1st stage | Initial denaturing time 5 min; annealing temperature 61°C; extension time 70 s |
| 355f-GC–516r (161 bp) | Eubacteria | DGGE/nested amplification 2nd stage | Initial denaturing time 2 min; annealing temperature 55°C; extension time 30 s; final extension time 5 min |
Table 1: Primers and PCR conditions.
Denaturing gradient gel electrophoresis
A Bio -Red D Gene TM system (Bio-Red, USA) was used to perform denaturing gradient gel electrophoresis analysis as per the method of Griffiths et al. [23] Briefly, the polymerase chain reaction amplification samples loaded of 8% (w/v) polyacrylamide gels in 1 X TAE . The polyacrylamide gels were prepared with a denaturing gradient ranging from 45% to 60%. Overnight electrophoresis was carried out for 15 hours at 60°C and 40 V, and then the gels were plunged for 20 minutes in 200 μL 1X TAE II supplemented with nucleic acid gel stain. Afterwards, the pictures of the gels were captured with a UV transillumination table in combination with a video camera module.
Analysis of denaturing gradient gel electrophoresis
The obtained denaturing gradient gel electrophoresis blueprints were then analyzed by Help Bio Numerics software (version: 7.5, Belgium). This software define the different levels, surroundings elimination, marker assisted normalization, which contains compensating for intensity differences between the orbits, and assigning different bands in each lane. An array of facilities for densiometric curves the band design was computed based on the Pearson product-moment correlation coefficient and dendrograms were constructed using UPGMA bond. Separations of Relevant and non-relevant clusters were again splited by the cluster cut-off method. An analysis of moving window consisted of plot progression in time of the connection values between two consecutive analyze dates and are useful when analyzing bacterial community stabilities [24]. For sequence analysis intention, the required denaturing gradient gel electrophoresis band fragments were excised and cloned by polymerase chain reaction 2.1- TOPO cloning kit according to the manual instructions. DNA sequencing was performed by Bangalore Genei. DNA sequence analysis was carried out using BLAST server of the National Center for Biotechnology Information with The BLAST algorithm and specifically with the BLASTN program.
Cloning and sequencing of 16S rRNA gene fragments
amoA PCR products were ligated according to the manufacturer recommendations in the cloning vector pCR2.1 delivered with TOPO TA cloning. Nucleotide sequences were determined on both strands by the dideoxynucleotide Procedure [25] of cycle sequencing of the purified plasmid with the Thermo Sequenase Cycle Sequencing Kit and an infrared automated DNA sequencer under conditions recommended by the manufacturers. Dye-labelled M13 targeted sequencing primers were used. 16S rDNA PCR amplificates obtained from ammonia oxidizing bacteria pure cultures was sequenced directly using primers targeting conserved regions. The novel 16S rRNA sequences were added to an adjustment of about 16,000 homologous primary structures from bacteria using customization tools of ARB program package. Alignments were purified by visual inspection. Phylogenetic analyzes were performed using distance matrix, maximum-parsimony and maximum-likelihood methods by respective tools of ARB and PHYLIP program packages and fast DNA ml program [26]. The composition of the datasets varied with respect to the reference value sequences and adjustment positions included. Variability’s of the particularized positions alignment was determined using ARB package and was used as the criteria for removing or including variable positions for the phylogenetic analyzes.
Denaturing gradient gel electrophoresis screening
Denaturing gradient gel electrophoresis was based on Core et al. [27]. In essence were group-specific PCR products used as template in a polymerase chain reaction with an embedded Bacterial denaturing gradient gel electrophoresis polymerase chain reaction primer pairs (Table 2). Semi-nested polymerase chain reactions were used in conjunction with the cyanobacteria/chloroplasts, and the Bacteroidetes Betaproteobacteria (Table 2). Only one bacterial primer was available within the 16S rRNA gene fragments, was amplified by the primers specific for these phylogenetic groups. Polymerase chain reaction conditions and cycle Protocol for the nested polymerase chain reaction with denaturing gradient gel electrophoresis primer pairs were the same as those used for polymerase chain reactions with the groupspecific primer pair, except for Ats as shown in table 2. Denaturing gradient gel electrophoresis of the polymerase chain reaction products was performed on 8% (w/v) polyacrylamide gel with urea and formamide as denaturing. The denaturing gradients are varied with the denaturing gradient gel electrophoresis primers used for polymerase chain reaction but were generally between 40% and 60% (Table 2). Electrophoresis was performed in Tris-acetate EDTA buffer at 60 0C at constant voltage of 60 V to 18 hours. Subsequently, the gels were stained for 1 SYBR Gold nucleic acid gel stain for 45 min and rinsed in distilled water before the picture analysis on a Syngeneic GELDOC station. Individual denaturing gradient gel electrophoresis bands were excised from the gel and incubated overnight at 4°C in 30 ml of H2O. An aliquot (Ca 5 mL) were used in a polymerase chain reaction with the same primer set used for denaturing gradient gel electrophoresis (Table 2, but without the GC clamp attached to a primer) to amplify the insert. The nucleotide sequences of the 16S rRNA gene fragments were determined as described above.
| Primers utilized for group-specific PCR | Primers utilized for re-PCR for DGGEa | AT (semi-) nested PCR (1C) | Denaturing gradient used for DGGE (%) b |
|---|---|---|---|
| Alf28f/Alf684 | 341f-GC/518r | 56 | 40-60 |
| Beta360f/Beta680r | 518f-GC/Beta682r | 60 | 40-55 |
| Gamma392f/Gamma870r | 518f-GC/785r | 56 | 40-60 |
| CFB550f/CFB966r | CFB555f-GC/907r | 64 | 40-60 |
| CYA361f/CYA785r | 518f-GC/CYA785r | 56 | 40-55 |
| Plancto352f/Plancto920r | 518f-GC/907r | 60 | 40-60 |
| Firm350f/Firm814r | 518f-GC/785r | 56 | 40-60 |
| 9bfm/1512uRc | 341f-GC/518r | 56 | 40-60 |
Table 2: Nested polymerase chain reaction approach with denaturing gradient gel electrophoresis primers.
ARDRA screening of cloned 16S rRNA genes
Amplified rDNA restriction analysis [28] was used to characterize the diversity of the gene 16S rRNA in culture collections and clone libraries. One, or in some cases several representatives of Ardra pattern groups from each culture collection and clone library were selected for sequencing. Sequence data were analyzed by ARB software package. Dendrograms was reconstructed phylogenetic analysis. The frequency of the 16S rRNA gene was determined by phylotypes Ardra and subsequent sequencing was used for the analysis of diversity. Shannon index of diversity (H) was calculated according to the method of Hammer [29]. Dilution curves were interpolated with freeware program Analytical diluting 1.3. Coverage of clone libraries was estimated as previously described by Ho et al. [30].
Phylogenetic analysis
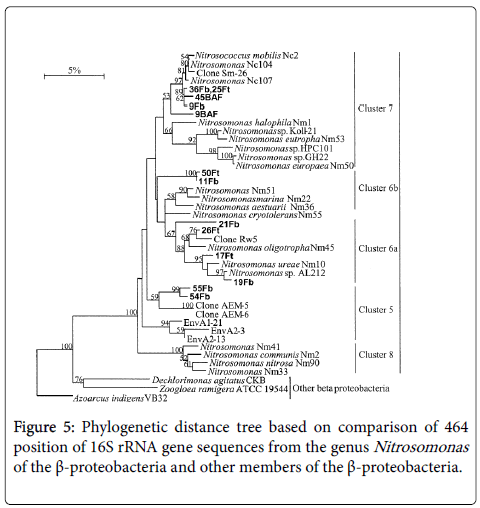
The incomplete 16S rRNA gene sequences from the reactor were adjusted to published 16S rRNA gene sequences from β- proteobacterial ammonia oxidizing bacteria and related non- ammonia oxidizing bacteria was used as out group sequences. A phylogenetic distance tree was generated using the Jukes and Cantor correction [31], and the neighbour - joining algorithm [32], implemented in TREECON software package.
DGGE pattern analysis
All data analyzes denaturing gradient gel electrophoresis profiles and environmental variables were performed using the software program Primer 6 [33]. The similarity of denaturing gradient gel electrophoresis profiles was determined using Bray-Curtis Index of similarity by the square root transformation. One similarity matrix made for all sites was used to make a dendogram using weighted-average group linkage in cluster analysis and nonmetric multidimensional scaling plots. Multidimensional scaling is ordination technique that is representative of the samples as points of a two-dimensional space. The relative distances between points in the same ranking as the relative similarities samples, i.e. points that are close to each other represent the samples that have very similar jointly composition. The procedure BIO-ENV in the software package PRIMER 6 was used to relate the main environmental factors to the bacterial community. This analysis consists of a similarity matrix obtained from denaturing gradient gel electrophoresis profiles for the resulting matrix of the Euclidean distances obtained after normalization of environmental data. This analysis allowed us to obtain the variable by largest correlations (weighted Spearman rank correlations). PRIMER 6 was also used to calculate species richness and Shannon index [34] from the denaturing gradient gel electrophoresis values.
Raup and Crick’s index
Wastewater treatment using an undefined complex community of microorganisms to treat various wastes. The assembly of these organisms to create a community sewage treatment plants is not well understood, but may be best seen in two ways. (1) Municipalities observations could be due to the selection of specific ¢ c organisms that are best adapted to the conditions in the race; Arises deterministic choice. (2) Municipalities may be present through the colonization of organisms present in the environment and specific organisms to colonize a wastewater treatment plant are determined by its chance arrival and plant propagation. Strictly test whether the similarities in the microbial community observed in comparing two sewage indeed the result of a deterministic selection, observed data should be statistically tested against the null hypothesis [35-38]. In this case, the null hypothesis is that the similarities can be measured can be explained by two organisms of the same wastewater. The biggest similarity is commonly used to compare the process DGGE is unable to recognize similarities due to random mating strips or adaptation strip at a level higher than would be expected by chance alone and statistically test the significance of the observed similarity against one null hypothesis is rarely explicitly stated in these comparisons. Method Raup and Crick [39] employs a random selection procedure for the detection of similarity greater than can be explained by coincidence the strips for two persons for ¢ forest and enables statistical significance similarities to be tested against a null hypothesis. Raup and Crick similarity index compares the number of species common to the two locations according to the number of species common to the two sites that could be expected if the species were chosen randomly from the source file. Differences in the observed data compared with those randomized to correlate with the level of similarity or dissimilarity between these two places.
Results and Discussion
A number of modern studies of ammonia oxidizing bacteria in wastewater treatment plants have recommended that different plants sustain different populations and diverse levels of species richness. For example, a household wastewater bio film from a lab - scale reactor was dominated by North Europe alike ammonia oxidizing bacteria [25] while the ammonia oxidizing bacterial populations from lab- and full scale was dominated by Nitroso spiral like bacteria or N. mobilis -like ammonia oxidizing bacteria respectively [12,27] . Polymerase chain reaction, denaturing gradient gel electrophoresis and sequence analysis was combined to determine the dominant ammonia oxidizing bacteria populations in biologically activated filter and trickling filter reactors.
Denaturing gradient gel electrophoresis
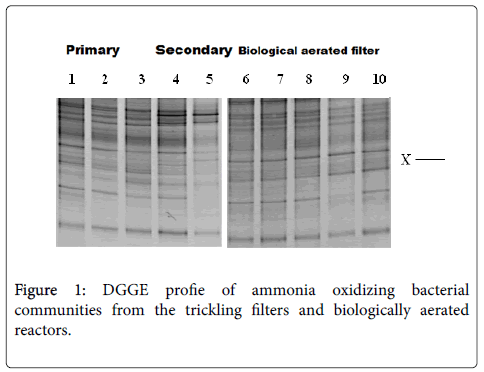
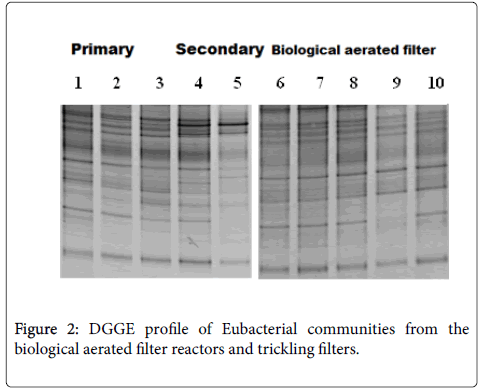
An exploration of duplicate samples by denaturing gradient gel electrophoresis showed that profiles gained were reproducible; comparisons of individual samples from each section of the reactor are shown. An optical assessment of denaturing gradient gel electrophoresis profiles by bacterial and ammonia oxidizing bacterial 16S rRNA gene fragments from filter beds and biological aerated filter reactor revealed some different populations of the different sections of each of the reactors and differences between the reactors (Figures 1 and 2). Bacterial and ammonia oxidizing bacterial denaturing gradient gel electrophoresis data for the biological aerated filter suggest the conditions of the reactor choose differentially different for some populations (Figures 1 and 2, lane 7-9). Biological Aerated Filter reactor consists of three connected pools each, which is optimized for different processes. An alteration in the plane of aeration seems to have an outcome on the bacterial-populations present. This is apparent by a visual assessment of the banding pattern of each of the three phases (Figures 1 and 2, lane 7-9). Differences in significant bacterial and ammonia oxidizing bacterial DGGE data was also scrutinized between the depths of different filter bed and between the primary and secondary filter bed (Figures 1 and 2). For example, several ribbons were exposed in samples from the bottom of the primary filter (Figure 1) and in the secondary filter bed (Figure 1) than at the top of the primary filter (Figure 1). Furthermore, particularly in the ammonia oxidizing bacterial denaturing gradient gel electrophoresis profiles, there seems to be a succession change in bacterial populations through filter beds. This is clearly the loss of some bands down filter bed pro ¢ le and the appearance of other (Figure 1).
An evaluation of DGGE profiles between filter biological aerated filter beds and the reactor showed that although both reactors were fed the same waste they harboured clear ammonia oxidizing bacteria and bacterial populations (Figures 1 and 2). Biological aerated filter reactor contained a lower detectable plurality of ammonia oxidizing bacteria compared to the filter beds. In particular, a number of bands that migrated further in the denaturing biologically aerated filter reactor. Yet both reactors seemed to have a common predominant population (marked X in Figure 1). To confirm identity ammonia oxidizing bacteria represented by bands denaturing gradient gel electrophoresis gels PCR - amplified 16S rDNA from waste water treatment plant samples were cloned and sequenced.
Ammonia oxidizing bacterial characterization
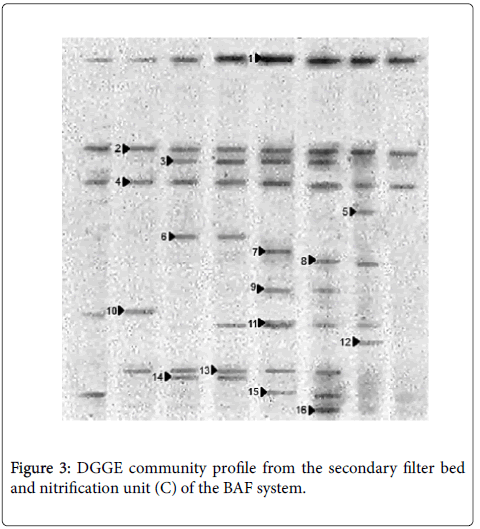
Clone Libraries of β-proteobacterial ammonia oxidizing bacterial 16S rRNA genes was assembled from samples from each of the reactors chosen as those that contain the greatest variety on the basis of denaturing gradient gel electrophoresis profiles or exhibiting active nitrification. Two methods denaturing gradient gel electrophoresis and ARDRA, was employed to screen 30 clones from each sample. Both methods resulted in similar groups of clones; However denaturing gradient gel electrophoresis confident more discriminatory than ARDRA, thus ARDRA appeared underestimating ammonia oxidizing bacterial diversity in these reactors. The denaturing gradient gel electrophoresis screening clone libraries available the sequences co-migrated with mainly of the prevailing bands from the novel denaturing gradient gel electrophoresis analysis was recovered in clone libraries (Figure 3). Conversely, some clones present at low frequencies is not in clone libraries have a similar band in denaturing gradient gel electrophoresis gel. Still, the intensity of the majority of the bands reflected different frequency of clones in the libraries. Nucleotide sequences were determined for each clone type from clone libraries and were compared with Gen Bank database [40-46]. All ammonia oxidizing bacterial sequences recovered was between 96 and 99% identity with the previously identified ammonia oxidizing bacterial 16S rRNA gene sequences with except for clones 21Fb and 55Fb which had 95% identity N. cryotolerans with Nm55 and environmental clone related to the Nitrosomonas sp. (Table 3).
| Clone type | Closest relative | Percent identity | Origin |
|---|---|---|---|
| 9BAF | Nitrosomonas sp. Nm107 | 98.4 | Activated sludge |
| 21Fb | Nitrosomonas crylotolerans Nm55 | 96.4 | Activated sludge |
| 26Ft | Clone Rw5 | 99.2 | Culture from activated sludge |
| 19Fb | Nitrosomonas sp. AL212 | 99.4 | Activated sludge |
| 17Ft | Nitrosomonas ureae Nm10 | 99 | Culture from activated sludge |
| 25Ft 36Fb 45BAF | Nitrosomonas sp. Nm107 | 98.4 | Culture from activated sludge |
| 11Fb 50Ft | Nitrosomonas sp. Nm51 | 98.4 | Activated sludge |
| 67Ft | Uncultured β-proteobacterium CRE-Fl40 | 98.2 | Activated sludge |
| 47Ft | Uncultured β-proteobacterium SBR1011 | 99 | Culture from activated sludge |
| 55Fb | Culture AEM-5 | 98.6 | Activated sludge |
| 43Ft | Uncultured β-proteobacterium 16S-8 | 99.2 | Culture from activated sludge |
| 40Fb | Uncultured bacterium Phos-He26 | 97.4 | Activated sludge |
| 1BAF | Uncultured γ-proteobacterium BioIuz K38 | 98.3 | Culture from activated sludge |
Table 3: Adjoining neighbour of the cloned sequences of the biologically activated filter reactor.
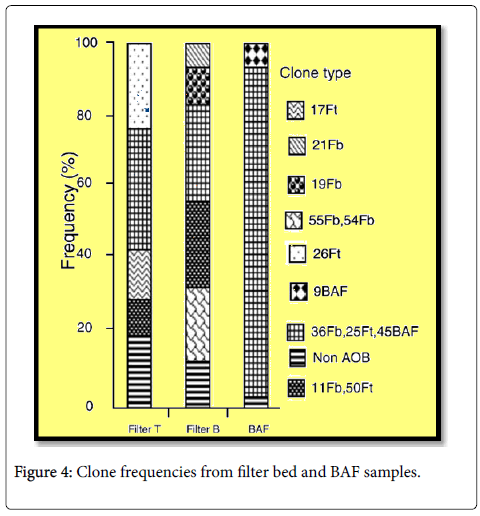
A depth analysis of the sequences recovered from biologically activated filter and filter bed revealed that all were derived from β- proteobacteria. Furthermore, most of the sequences were most closely related to β-proteobacterial ammonia oxidizers. The rest of the sequences recovered were usually closely related to either Thauera spp. or Dechlorimonas agitatus-like bacteria (Table 2). Ammonia oxidizing bacteria -like sequences from Nitrification unit of the BAF and the top and bottom of the filter bed reactors was all recovered, with the genus Nitrosomonas (Figure 4). The BAF clone library from the reactor had preponderance N. mobilis-like sequences (Figure 4 and Table 3) (Nitrosomonas cluster 7). Whereas filter bed samples were more diverse, and three or four different predominant clone was identified types bed; N. freedoms like sequences Nitrosomonas spp. 6b cluster sequences, cluster sequences 6a and Nitrosomonas spp. Cluster 5 sequences Figure 4 and Table 3). The sequences most closely related to the cluster 6a, 6b and 5 had relatively low sequence identity to the most similar sequences in the sequence databases and their connection with these groups based on bootstrap analysis was not robust. This may be a consequence of the relatively short sequences used in the analysis. Analysis of long-16S rRNA gene sequences would help place these sequences con more ¢ arthritis with one of recognized ammonia oxidizing bacteria clusters. The nearest neighbours sequences reported were originally recovered from a range of environments but a number of group sequences from halo tolerant ammonia oxidizing bacteria or sequences recovered from salt water environments, or enrichments. N. mobilis-like sequences were found in all the reactors, but were relatively more abundant in the clone library from BAF (90% of sequences) than in filter beds (33% on top of the secondary, 27% at the bottom of secondary, Figure 4 and Table 3). It should be noted that AOB-selective primers that were used in this study, do not completely universal for all β-proteobacterial Optionally and primer CTO189f and CTO654r have mismatches with Some sequences from Nitrosomonas cluster and the cluster 7 6a and CTO654r have more than two mismatches with all cultured members of N. communis cluster (cluster 8) [47]. This may to some extent explain why we did not do detect any sequences closely related to Nitrosomonas cluster 8, while we detecting sequences related to all other currently described Nitrosomonas clusters, albeit in some cases of relatively low sequence identity. However, in clone libraries generated from some of the samples analyzed in this study, using the primers and Nso190 Nso1225, which has no differences with N. communis cluster [47], were not members of this group discovered (unpublished data). Cloning and sequencing analysis showed that comparable levels of AOB diversity were identified when amplified gene fragmenter rRNA was analyzed by DGGE or cloning. DGGE and sequencing analysis therefore seemed to indicate that the particular AOB were selected for the different the reactors. To examine whether the selection of and in particular in the AOB reactors were statistically different significant Raup and Crick simulations [48-50] was employed.
Statistical analysis
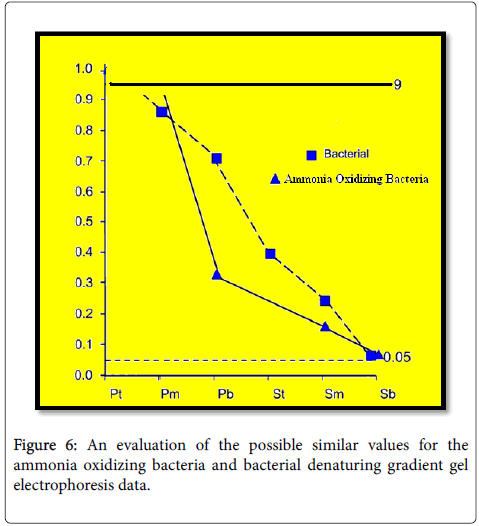
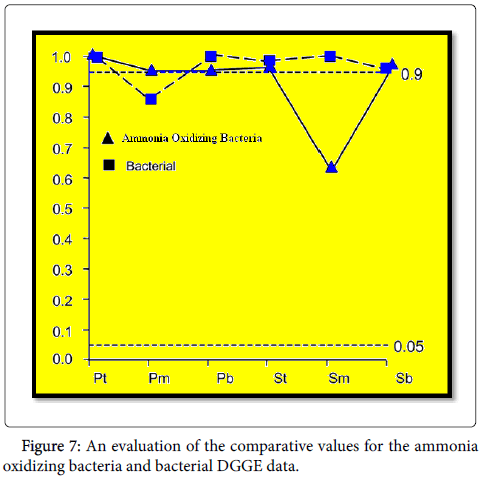
In our study, we detected N. mobilis-like organisms as a important component of ammonia oxidizing bacteria community in all samples studied. It is tempting to impose an interpretation when these patterns are observed. For example, saline wastewater promotes the occurrence of N. mobilis. However, it is inappropriate to interpret organic data before we have found that the patterns we observe cannot be explained by chance alone [51]. It is therefore necessary first to show that the patterns we observe cannot attributed to random distribution of species before derive deeper causal relationships between environmental and physiological factors and the incidence of specific organisms. Therefore, we tested the null hypothesis the similarity between patterns observed in DGGE data from our samples could be explained different by chance alone (Figures 5-7).
Ammonia oxidizing bacterial communities comparison
The similarity between the overall pooled ammonia oxidizing bacteria data and any given sampling point in rippling filter was no greater than could be explained by chance alone (P=0.501). There was also no significant equality between the top of the reactor and the subsequent depths (Figure 5). These data suggest that similarities in the ammonia oxidizing bacteria community present at different depth are not greater than can be explained by chance. But regression analysis showed a statistically significant (P=0.006) relationship between depth in the reactor and the SRC value. In addition, adjacent sampling points were similar (SRC>0.95; Figure 5) suggests that the choice of different Possibly with depth in filter was nonrandom and Similarities in ammonia oxidizing bacteria community was statistically greater than would be expected from chance matching of bands The DGGE profiles. There is clearly a significant gradient in ammonia oxidizing bacteria diversity into the reactor. A similar pattern of succession was also observed in the bacterial production DGGE profiles (Figures 5 and 6). This may be due to the stratification exist in neat bio film reactors that are waste degraded and changes in composition as it passes through the filter bed [40]. Waste enters the primary filter bed will contain a large amount of organic material, and waste degrades the level of organic matter will be reduced while oxygen will be consumed. You presence of high levels of organic matter can be heterotrophs outcompete autotrophic ammonia oxidizing oxygen [41] and ammonia [52,53]. As organic matter decreases, the number of heterotrophs, and thus decreases ammonia oxidizing bacteria can proliferate. There should therefore be a higher number of ammonia oxidizing bacteria in the bottom of the primary filter bed, and in the secondary filter bed. Possibly was not quantified in this study, but operational data suggest that secondary filter beds remove more ammonia than the primary Beds; shortly before the primary sampling 74%; Secondary 81%), suggesting the presence of a greater number of more or metabolic active AOB in the secondary filter. Interestingly, no statistically pursuit differences or gradients were observed for BAF although this is also a set of bio film reactor. This may be, at least partially, attributed to the more uniform conditions spawned the strong mix and reuse regime BAF reactor. Comparison of DGGE data from BAF and rippling filter showed that for all but two of the reactor comparisons similarities observed was not greater than would expected by random association of bands in DGGE process (Figure 7). Therefore, any differences observed between BAF and filter beds could be explained by random processes without relying on differences in reactor design as a causal factor. Nevertheless, the similarity values for each comparison, BAF with the primary filter bed was generally all very high, while the values for each comparison, biologically activated filter with the secondary filter bed was lower and successively decreased with depth (Figure 7). Even though not statistically significant on the basis of the Raup and Crick analysis, it is clear that the similarities between the biologically activated filter and the primary filter bed are considerably greater than the similarities between the biologically activated filter and Secondary filter bed. Interestingly, evidence reports from providers that filter beds are more robust from Nitrification failure than other systems [54]. The comparison of the DGGE pooled data from biologically activated filter with the secondary filter bed suggested that ammonia oxidizing bacteria populations biologically activated filter and secondary filter bed was dated ¢ known from diverging. This was reflected in the level of ammonia oxidizing bacteria diversity within each plant. The filter beds had greater ammonia oxidizing bacteria diversity than the biologically activated filter. This could explain why filter beds doing better and are inherently more stable than biologically activated filter reactor. It has been suggested that the level of diversity ammonia oxidizing bacteria in a sewage treatment plant has great influence on the process stability [55]; the greater the diversity, the more stable process. A plant with greater diversity, should cope better changing conditions, since a reduction in the number of one organism may not mean the process failure as other organisms better adapted to the new conditions can multiply resulting in a more functionally stable system. Our data support this view as filter beds where ammonia oxidizing bacteria diversity was bigger; they are inherently more stable reactors. If diversity does play a big role in wastewater treatment then as before by the demands of and his staff proposed [56], the design of the plant to have higher diversity making processes as nitrification more stable. However, the reason different ammonia oxidizing bacteria populations are present having different conditions shall be understood to allow the development of technical solutions to obtain most suitable diverse ammonia oxidizing bacteria population purifiers. The observation of gradients in diversity of ammonia oxidizing bacterial communities suggests that the environment is the selection partly influencing diversity observed. However, N. mobilis-like organism seems to be ubiquitous on this site. It means that either this organism is well adapted to the environmental conditions in all places at all reactors or is so rich source community, as demonstrated by coincidence at all locations [45]. Traditional microbial ecologists have taken more deterministic approach [57-66]. But we cannot distinguish between these two possibilities in this or any other, purely observational studies[67-69]. This is probably a number of mechanisms at work, and the relative importance of each mechanism will be different. In our study, for example, that N. mobilis as dominant ammonia oxidizing bacteria at all depths ¢ filter bed in the event that the assembly community was random and N. mobilis like ammonia oxidizing bacteria were abundant in the source population. In fact, other ammonia oxidizing bacteria sequences increase proportional to the deeper into the reactor, observations in accordance with a deterministic selection. Reactors are studied, being fed effluent from the same HRAS reactor. Interestingly, although there are no reports on the composition of the ammonia oxidizing bacteria communities in the reactor effluent HRAS, analysis of the cloned gene sequence of 16S rRNA derived from this reactor by means of ammonia oxidizing bacteria -selective primers showed that the clone libraries were low rate of ammonia-oxidizing bacteria sequence (26%, n=30), but those that were identified were most closely related to N. mobilis (data not shown). In this study, we have shown important and significant differences in the ammonia oxidizing bacteria diversity two complete wastewater treatment different conformations. Differences in diversity may very well be related to differences in performance. The challenge now is to elucidate the mechanism underlying the differences so that these systems can be integrated into the design of the waste water treatment plant.
References
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, et al. (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102-112.
- Green JL, Bohannan BJ, Whitaker RJ (2008) Microbial biogeography: from taxonomy to traits. Science 320: 1039-1043.
- Horner-Devine MC, Lage M, Hughes JB, Bohannan BJ (2004) A taxa-area relationship for bacteria. Nature 432: 750-753.
- Kowalchuk GA, Stephen JR (2001) Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol 55: 485-529.
- Purkhold U, Wagner M, Timmermann G, Pommerening-Roser A, Koops HP (2003) 16 S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the Nitrosomonas. Int J Syst Evol Microbiol 53:1485-1494.
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, et al. (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806-809.
- Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10: 2931-2941.
- Webster G, Embley TM, Freitag TE, Smith Z, Prosser JI (2005) Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ Microbiol 7: 676-684.
- Boyle-Yarwood SA, Bottomley PJ, Myrold DD (2008) Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ Microbiol 10: 2956-2965.
- Carney KM, Matson PA, Bohannan BJM (2004) Diversity and composition of tropical soil nitrifiers across a plant diversity gradient and among land-use types. Ecol Lett 7: 684-694.
- Mintie AT, Heichen RS, Cromack K Jr, Myrold DD, Bottomley PJ (2003) Ammonia-oxidizing bacteria along meadow-to-forest transects in the Oregon Cascade Mountains. Appl Environ Microbiol 69: 3129-3136.
- Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ (2008) Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10: 1601-1611.
- Avrahami S, Liesack W, Conrad R (2003) Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol 5: 691-705.
- Horz HP, Barbrook A, Field CB, Bohannan BJ (2004) Ammonia-oxidizing bacteria respond to multifactorial global change. Proc Natl Acad Sci U S A 101: 15136-15141.
- Kowalchuk GA, Stienstra AW, Heilig GHJ, Stephen JR, Woldendorp JW (2000) Changes in the community structure of ammonia oxidizing bacteria during secondary succession of calcareous grasslands. Environ Microbiol 2: 99-110.
- Chu H, Fujii T, Morimoto S, Lin X, Yagi K, et al. (2007) Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Appl Environ Microbiol 73: 485-491.
- Bottomley PJ, Taylor AE, Boyle SA, McMahon SK, Rich JJ, Cromack K, Myrold DD (2004) Responses of nitrification and ammonia-oxidizing bacteria to reciprocal transfers of soil between adjacent coniferous forest and meadow vegetation in the Cascade Mountains of Oregon. Microbial Ecol 48: 500-508.
- Avrahami S, Conrad R (2005) Cold-temperate climate: a factor for selection of ammonia oxidizers in upland soil? Can J Microbiol 51: 709-714.
- Kowalchuk GA, Stephen JR, De Boer W, Prosser JI, Embley TM et al. (1997) Analysis of ammonia-oxidizing bacteria of the L subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol 63: 1489-1497.
- Harris SJ, Mortimer RJG (2002) Determination of nitrate in small volume samples (100 µl) by the cadmium-copper reduction method: a manual technique with application to the interstitial waters of marine sediments. Int J Environ Anal Chem 82: 369-376.
- Hayes P (2001) Diagenetic processes and metal mobilisation in an organic rich Scottish fjord. Department of Earth Sciences, University of Leeds, Leeds.
- Nicol GW, Glover LA, Prosser JI (2003) The impact of grassland management on archaeal community structure in upland pasture rhizosphere soil. Environ Microbiol 5: 152-162.
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ (2000) Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66: 5488-5491.
- Albertsen M, Hansen LB, Saunders AM, Nielsen PH, Nielsen KL (2012) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6: 1094-1106.
- Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacher M, et al., (2007) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 36: D623-D631.
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, et al. (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141-145.
- Dubbels BL, Sayavedra Soto LA, Bottomley PJ, Arp DJ (2009) Thauera butanivorans sp. Nov., a C2-C9 alkane-oxidizing bacterium previously referred to as ‘Pseudomonas butanovora’. Int J Syst. Evol Microbiol 59: 1576-1578.
- Foss S, Harder J (1998) Thauera linaloolentis sp. nov. and Thauera terpenica sp. nov., isolated on oxygen-containing monoterpenes (linalool, menthol, and eucalyptol) nitrate. Syst Appl Microbiol 21: 365-373.
- Hammer O, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electro. 4: 1-9.
- Ho CM, Tseng SK, Chang YJ (2001) Autotrophic denitrification via a novel membrane-attached biofilm reactor. Lett Appl Microbiol 33: 201-205.
- Hu M, Wan, X, Wen X, Xia Y (2012) Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol 117: 72-79.
- Kurt M, Dunn IJ, Bourne JR (1987) Biological denitrification of drinking water using autotrophic organisms with H(2) in a fluidized-bed biofilm reactor. Biotechnol Bioeng 29: 493-501.
- Liessens J, Vanbrabant J, De Vos P, Kersters K, Verstraete W (1992) Mixed culture hydrogenotrophic nitrate reduction in drinking water. Microb Ecol 24: 271-290.
- Macy JM, Rech S, Auling G, Dorsch M, Stackebrandt E, et al. (1993) Thauera selenatis gen. nov., sp. nov., a member of the beta subclass of Proteobacteria with a novel type of anaerobic respiration. Int J Syst Bacteriol 43: 135-142.
- Madden TL, Tatusov RL, Zhang J (1996) Applications of network BLAST server. Methods Enzymol 266: 131-141.
- Maldonado LA, Fragoso-Yáñez D, Pérez-García A, Rosellón-Druker J, Quintana ET (2009) Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 95: 111-120.
- Mansell BO, Schroeder ED (2002) Hydrogenotrophic denitrification in a microporous membrane bioreactor. Water Res 36: 4683-4690.
- Mateju V, Cizinska S, Krejci J, Janoch T (1992) Biological water denitrification-a review. Enzyme Microb Technol 14: 170-183.
- Mechichi T, Stackebrandt E, Gad'on N, Fuchs G (2002) Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromaticasp. nov., and Azoarcus buckelii sp. nov. Arch Microbiol 178: 26-35.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537-7541.
- Scholten E, Lukow T, Auling G, Kroppenstedt RM, Rainey FA, et al. (1999) Thauera mechernichensis sp. nov., an aerobic denitrifier from a leachate treatment plant. Int J Syst Bacteriol 3: 1045-1051.
- Smith RL, Buckwalter SP, Repert DA, Miller DN (2005) Small-scale, hydrogenoxidizing- denitrifying bioreactor for treatment of nitrate-contaminated drinking water. Water Res 39: 2014-2023.
- Song B, Palleroni NJ, Kerkhof LJ, Häggblom MM (2001) Characterization of halobenzoate-degrading, denitrifying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int J Syst Evol Microbiol 51: 589-602.
- Sunger N, Bose P (2009) Autotrophic denitrification using hydrogen generated from metallic iron corrosion. Bioresour Technol 100: 4077-4082.
- Szekeres S, Kiss I, Kalman M, Soares MI (2002) Microbial population in a hydrogen-dependent denitrification reactor. Water Res 36: 4088-4094.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731-2739.
- Tang Y, Zhou C, Ziv-El M, Rittmann BE (2011) A pH-control model for heterotrophic and hydrogen-based autotrophic denitrification. Water Res 45: 232-240.
- Till BA, Weathers LJ, Alvarez PJJ (1998) Fe(0)-supported autotrophic denitrification. Environ Sci Technol 32: 634-639.
- Vasiliadou IA, Pavlou S, Vayenas DV (2006) A kinetic study of hydrogenotrophic denitrification. Process Biochem 41: 1401-1408.
- Vasiliadou IA, Siozios S, Papadas IT, Bourtzis K, Pavlou S, et al. (2006) Kinetics of pure cultures of hydrogen-oxidizing denitrifying bacteria and modeling of the interactions among them in mixed cultures. Biotechnol Bioeng 95: 513-525.
- Lee KC, Rittmann BE (2003) Effects of pH and precipitation on autohydrogenotrophic denitrification using the hollow-fiber membrane- biofilm reactor. Water Res 37: 1551-1556.
- Sayess RR, Saikaly PE, Fadel ME, LiD, Semerjian L (2013) Reactor performance in terms of COD and nitrogen removal and bacterial community structure of a three-stage rotating bioelectrochemical contactor. Water Res 47: 881-894.
- Bastiaens L, Springael D, Wattiau P, Harms H, deWachter R, et al. (2000) Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl Environ Microbiol 66: 1834-1843.
- Boldrin B, Tiehm A, Fritzsche C (1993) Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol 59: 1927-1930.
- Laura AS, Francien P, Stefan S, Josh DN (2012) Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ Microbiol 14: 2589-2600.
- BjO¨ rnsson L, Hugenholtz P, Tyson GW, Blackall LL (2002) Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148: 2309-2318.
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, et al. (2010) Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci U S A 107: 1612-1617.
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science 324: 1190-1192.
- van der Wielen PW, Voost S, van der Kooij D (2009) Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol 75: 4687-4695.
- Mao Y, Yannarell AC, Davis SC, Mackie RI (2013) Impact of different bioenergy crops on N-cycling bacterial and archaeal communities in soil. Environ Microbiol 15: 928-942.
- Morris RM, Nunn BL, Frazar C, Goodlett DR, Ting YS, et al. (2010) Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J 4: 673-685.
- Barberán A, Bates ST, Casamayor EO, Fierer N (2012) Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343-351.
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA (2006) Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532-2538.
- Ju F, Xia Y, Guo F, Wang ZP, Zhang T (2014) Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16: 2421-2432.
- Newman ME (2006) Modularity and community structure in networks. Proc Natl Acad Sci U S A 103: 8577-8582.
- Croft DP, Krause J, James R (2004) Social networks in the guppy (Poecilia reticulata). Proc Biol Sci 271 Suppl 6: S516-519.
- Shuttleworth KL, Unz RF (1993) Sorption of heavy metals to the filamentous bacterium thiothrix strain A1. Appl Environ Microbiol 59: 1274-1282.
- Wrighton KC, Virdis B, Clauwaert P, Read ST, Daly RA, et al. (2010) Bacterial community structure corresponds to performance during cathodic nitrate reduction. ISME J 4: 1443-1455.
- Castignetti D, Hollocher TC (1984) Heterotrophic nitrification among denitrifiers. Appl Environ Microbiol 47: 620-623.
Copyright: © 2016 Shah MP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.