Fisheries and Aquaculture Journal
Open Access
ISSN: 2150-3508
ISSN: 2150-3508
Research Article - (2021)
A microsatellite DNA marker technique was used to monitor specific-pathogen-free Penaeus vannamei (white shrimp) stock in the hatchery of the University of Guam, mainly for assessment of genetic diversity and identification of the parentage for two consecutive generations. A panel of 16 loci was selected to analyze 36 families of P. vannamei, comprising of a total of 1,152 individual shrimp samples. The families showed high genetic variation. The average number of alleles per locus was 10.625 for the parents and 10.052 for their offspring. The average observed heterozygosity declined slightly from 0.891 in the parents to 0.813 in the offspring. Similarly the expected heterozygosity was 0.804 among the parents and 0.792 for their progeny. The inbreeding coefficient was -0.107 for the parents but -0.026 for the offspring. These two generations did not differ significantly in any foregoing measures (p>0.05). CERVUS and COLONY confirmed that any 12 loci from the panel could deliver 100% correct parentage identification when the genotyping error rate was set as 0.01.
Genetic diversity; Selective breeding program; Penaeus vannamei; Microsatellite; Inbreeding
Shrimp aquaculture has boomed in the past fifty years, and Penaeus vannamei Boone, synonym Litopenaeus vannamei) stands out as the dominant penaeid species cultivated since 2005, and has become the predominant crustacean in the world aquaculture. Being the first penaeid species to which the specific-pathogen-free (SPF) concept was applied, combined with substantial efforts of selective breeding with a history about 30 years, are the two major c factors contributing to P. vannamei’s dominance in the market. Shrimp disease outbreaks are the major obstacle to of aquaculture development. The initiation of the first breeding attempt for developing a shrimp stock free of infectious hypodermal and hematopoietic necrosis virus (IHHNV), the domesticated SPF P. vannamei population, took place from 1990 to 1991. Several years later, taura syndrome virus (TSV) broke out and led directly to more catastrophic loss in shrimp production than IHHNV. The domesticated populations were therefore selected for TSV resistance in addition and also bred for better growth and survival rate in various culture systems, including both zero water exchange system and brackish-water environments. The penaeid disease list of the World Organization for Animal Health (OIE) and the working list of U.S. Marine Shrimp Farming Program (USMSFP) are the two sources most commonly referred to for establishing SPF status, but the active list of SPF strains has expanded as the known shrimp pathogens of economic significance have emerged over time. The list of viral pathogens increased from those focused on early IHHNV, TSV, white spot syndrome virus, yellow head virus to include infectious myonecrosis virus and other types of pathogens. Bacterial diseases were necrotizing hepatopancreatis and acute hepatopancreas necrosis disease. More recently, hepatopancreatic microsporidiosis caused by Enterocytozoon hepatopenaei spurred great concern, even though it is not yet included in OIE disease list yet. So far, worldwide, several genetic programs supported either by governments or by the private sectors provide of SPF P. vannamei broodstock or seed stock for local hatchery/grow out farm needs or the international market. In these breeding programs, shrimp stock with a highly diversified genetic background would help in the selection for any heritable phenotypic traits among the population [1-16].
Abrupt reduction of genetic diversity could commonly result from using a narrow genetic pool and a small quantity of broodstock, as well as from inadvertent selection. As selection intensity increases, genetic variance inevitably declines over time in all domesticated breeding programs, but such reduction should be kept to a minimum by proper shrimp stock management. Without systematic mating design and careful execution, close relatives could breed, leading to significant inbreeding depression within only several generations. Inbreeding leads to two undesirable effects. It increases the probability of recessive allele expression and it can cause poor production or reproductive capacity, increased susceptibility to diseases, and other undesiable phenotypic traits. Selecting the genetically differentiated broodstock for mating with explicit pedigree information is vital control of inbreeding, for broodstock management in breeding practices, and for sustainability of shrimp breeding programs. Molecular techniques using genetic markers are promising tools for accomplishing this task. Among various genetic markers, the abundant presence of highly polymorphic microsatellites proves to be particularly useful in analyzing population variance, identifying the pedigree relationship in the breeding program as they exhibit Mendelian inheritance and are codominantly expressed. For P. vannamei, microsatellite markers can be developed from DNA sequences in the genome database. Markers developed for various penaeid shrimp species were applicable in studying the population genetics, for example, determining genetic relationships between the populations evaluating genetic diversity of the breeding stocks, and generating the linkage maps. In addition, these microsatellite loci could assist in pedigree identification. Establishment of a panel of informative microsatellite loci with the proper characteristics and number is imperatively important for preventing inbreeding in shrimp stock management. At any acceptable rate of accuracy in parental assignment, genotyping with the minimal number of loci adequate to link any shrimp correctly with its parents would make economic sense. The SPF P. vannamei breeding program has been housed at the biosecured Guam Aquaculture Development and Training Center (GADTC) since 2007, and has been under specific and general surveillance and tested routinely for all known significant penaeid diseases by the aquaculture pathology lab at the University of Arizona, an OIE referred lab. The SPF stock is free of OIE and USMSFP-listed shrimp pathogens, as well as other emerging pathogens, acute hepatopancreas necrosis disease, Baculovirus penaei, Enterocytozoon hepatopenaei, hepatopancreatic parvovirus, IHHNV, infectious myonecrosis virus, necrotizing hepatopancreatis, TSV, white spot syndrome virus, and yellow head virus. By applying the microsatellite markers, we sought (1) to monitor genetic diversity and inbreeding index in two consecutive generations in Guam’s breeding program and (2) to assign parentage correctly to offspring on the basis of genotyping results from a panel of selected microsatellite markers [11,12,17- 39].
Generating shrimp families
From the SPF brood stocks in Guam’s breeding program, 36 male-female pairs were selected to generate 36 offspring families by the means of artificial insemination (AI), according to the mating matrix design constructed on the basis of their genetic background and their familial performance data. After insemination, each mated female was held to spawn in a covered 1000 liters clean seawater tank equipped with adequate levels of aeration, temperature (28°C), salinity (34 ppt), and pH (7.4). Each female was gently netted and removed from the tank after spawning, while the fertilization of gametes and hatching process took place within the same tank. Once successfully hatched, approximately 3000 nau plii from each spawning tank were collected within 36 hours by phototaxis, cleaned, and then transferred to and reared in an individual larval rearing tank (38 L) with temperature is controlled at 30°C to 32°C. Water exchange was conducted as necessary to maintain water quality desirable for larval development. A variety of larval feeds (microalgae, live Artemia, artificial feed) of proper size and the adjusted amount was administered every four hours to ensure optimal nutrition for each of three distinct larval stages (nauplii, zoea, and mysis) before larvae reached the postlarval stage. During this stage, the daily water exchange rate increased from 30% to 100% in each tank. On the 12th postlarval day, each family was moved to early grow-out tanks (1,000 L) for faster growth. These postlarvae were fed four times daily with mixed artificial feed and live Artemia until they reached 2 to 3 grams, concluding the generation of one family of shrimp. About 300 shrimp from each family underwent physical tagging with visible implant elastomer (VIE) and were grown outdoors until ready for sampling. Different combinations of color-coded elastomer at two sites out of three locations in the muscle of the sixth abdominal segment served to identify each family uniquely.
DNA sampling and extraction
A pair of pleopods was taken from each of the 30 randomly selected individual shrimp from each of the 36 families produced and their parents. The specimens were kept frozen at -80°C until used for genomic DNA extraction. There were 1,152 shrimp specimens, comprising samples from 72 parents and 1,080 offspring derived from the 36 families, were all submitted for DNA extraction. The DNA extraction kit for cell/blood (Bioteke Co. Ltd., Beijing, China) was used for genomic nucleotide extraction according to the manufacturer’s protocols provided. Concentration of extracted DNA was measured, and the samples were kept at -4°C until further genotyping analysis.
Microsatellite DNA marker analysis
The DNA extracted from each individual shrimp was genotyped with all 16 informative microsatellite markers.To form this panel of 16 useful loci, we prescreened 128 microsatellite loci developed by previous studies, and tested among the pooled GADTC shrimp stocks for their highly polymorphic allelic presence in Guam’s population. Details of these 16 highly polymorphic microsatellite loci, such as source, size of various amplicons, and annealing temperature for each locus are provided in Table 1.
| Locus | Primer sequences (5′→3′) | Core sequence | Size range (bp) | Ta (°C) | Gen bank no |
|---|---|---|---|---|---|
| TUGAPv 7-9.119 | F:CATGACCTGCCTTTAATCCC | …(TC)15…(CT)8…(C)10(CT)5A(TC)3TT(TC)8A( | 313 - 389 | 52 | AY376983 |
| R:AAAGACAAGGAACGAGCGAG | CT)9…(CT)5…(TC)4…(CT)5...(CT)3…(TC)3… | ||||
| TUMXLv 5.66 | F:GGGGCACTGAGACGAGTAAG | …(CA)4CG(CA4T(AG)3…(TA)3…(AC)3…(AC)4 | 243 - 329 | 50 | AF360027 |
| R:CCGTTTTATCAGTCTCCATATACGA | …(AC)6… | ||||
| TUMXLv 7.56 | F:CCATGGCTTTCCTCTTCTTTC | …(TCC)5…(CCT)3…(CCT)3…(TC)4…(TC)4…( | 277 - 561 | 54 | AF360055 |
| R:AGGTAGGGAAGTCGTGAGGG | TC)3…(TC)3…(TC)3…(CT)3…(CCT)3… | ||||
| TUMXLv 7.97 | F:TGTCGTTAGTGCAGCTCATTC | …(AT)5…(TTC)3TT(TTC)3TT(TTC)3…(TCC)3C | 154 - 284 | 54 | AF360057 |
| R:GGGGAGGAATAAGAGGAAAGG | (TCC)5 | ||||
| TUMXLv 7.121 | F:GGCACACTGTTTAGTCCTCG | (GTT)3…(GA)3…(TC)3…(GT)3…(TC)3…(TC)3 | 206 - 307 | 52 | AF360043 |
| R:CGAACAGAATGGCAGAGGAG | …(TC)3… | ||||
| TUMXLv 7.138 | F:AGACACATACAGACGCACGC | …(CA)6TAG(AC)3GC(AC)3…(AC)3…(CA)19…( | 295 - 508 | 50 | AF360048 |
| R:GAGTTGCTCCCAAACGCTAC | CA)7… | ||||
| TUMXLv 8.193 | F:GATGTACACAACTGTACTTC | …(AT)3… | 188 - 235 | 53 | AF360069 |
| R:GAGATGATAAGAGAACGAAAG | |||||
| TUMXLv 8.256 | F:GGACTCACACTTCTGGTTC | …(AAT)4… | 166 - 207 | 53 | AF360076 |
| R:GGCTGCACCTTGTAAGTC | |||||
| TUMXLv 9.93 | F:CACCACCGAAAAGGTAGGAG | …(TG)5ATG(GT)4…(AG)9… | 306 - 428 | 50 | AF360115 |
| R:TGGGAGAGGTTAGTCATGGG | |||||
| TUMXLv 9.116 | F:GATGACCTGCCTTTCTCTGC | …(TTC)3…(CTT)3…(CT)3…(CTT)3…(GT)3T(T | 124 - 204 | 54 | AF360094 |
| R:GGGAGAGATGATGGGAAGAAG | CC)3…(TCC)3…(CT)3… | ||||
| TUMXLv 9.178 | F:CATTGAAAACGGAATCCTCG | …(GC)3…(CT)5… | 214 - 261 | 53 | AF360105 |
| R:GATATTCCCATCAACACAGCG | |||||
| TUMXLv 10.14 | F:CAGTCTACACGCACAGGCAC | …(CA)35…(CA)3…(CCG)3… | 216 - 304 | 53 | AF359947 |
| R:TTATACGGCGGTTCTCTTGG | |||||
| TUMXLv 10.33 | F:CGAAGAGATTTATCCAGGG | …(TTC)3…(ATA)3…(AAT)3…(AAT)4…(GA)3 | 201 - 388 | 53 | AF359992 |
| R:CGTGCATTATTATCCTTTCC | …(ATC)3… | ||||
| TUMXLv 10.341 | F:CATATGTATCTGCCCTCGAC | …(TCCC)3…(CCT)3…(CT)3…(CCCT)3… | 190 - 285 | 52 | AF359995 |
| R:TAGGATGGTGGGTAGAGTTG | |||||
| TUMXLv 10.411 | F:AGCACCTAGCACTTGCTGAAC | …(TC)3…(AAT)3…(TAA)12C(AAT)4… | 169 - 215 | 52 | AF360004 |
| R:AGAGACTCACATCCTCATCCTC | |||||
| TUMXLv 10.455 | F:AGAGTAGAAGAGGCAGGGCG | …(CT)7…(CT)10…(TC)5CC(TC)C(CT)3…(CT)4 | 257 - 386 | 50 | AF360007 |
| R:GTCAAGAAGCAGGAAGGGTG | TT(CT)CG(CT)CC(CT)5…(CT)3… |
Table 1: Detailed information of the 16 P. vannamei specific microsatellite loci being selected for this present study.
The total 12 μL PCR reaction reagents contained 1X Taq PCR MasterMix (Tiangen Biotech Co. Ltd., Beijing China), 50 ng DNA from each shrimp, and 0.42 ng each of the locus specific primer sets, forward and reverse. PCR products were amplified in the Labnet Multigene Thermocycler (Labnet International, Inc., USA), according to the following protocol: initial denaturation at 94°C for 3 min; 34 cycles of 94°C denaturing for 30s, the locusspecific annealing temperature for 30 s, and 72°C extension for 40 s; and a final extension at 72°C for 5 min. PCR product in the amount of 3.5 μL was loaded in 9.5% denaturing polyacrylamide gel and run at 200 volts for 135 min for allele separation, along with the standard marker, pBR322 DNA/MspI ladder (Tiangen Biotech Co.Ltd., Beijing, China) for comparison on each gel for the electrophoresis. For each PCR run, a negative control was included that contained all the remaining PCR reaction reagents other than the DNA template. After each electrophoretic run, 0.1% silver staining was applied to the gel to make the separated DNA fragments visible, followed by the gel’s image was captured with a Bio-Rad Imager (Bio-Rad Laboratories Ltd., USA). Genotype recording and data analysis were subsequently processed [28,31].
Genetic variability analysis
Multiple software packages were used for various genetic diversity analyses. First, Bio-Rad Quantity One version 4.6.2 software (Bio-Rad Laboratories Ltd., USA) was employed to extract quantitative information about alleles on the basis of gel genotypic recording. Second, Microsatellite Toolkit was applied to calculate allele frequencies, the number of alleles per locus (Na), observed heterozygosity (Ho), and expected heterozygosity (He) of each microsatellite locus, in different groups. Third, FSTAT 2.9.3 was used to obtain the inbreeding coefficient (Fis) in each of the two generations and population pairwise fixation index (Fst). Finally, for study of genetic differentiation among offspring families, PCAGEN 1.2.1 was used to conduct principal component analysis [40-42].
Pedigree analysis
Parentage assignment simulation involved two types of software packages, COLONY and CERVUS 3.0, and genotypic data generated from 30 individual shrimp from each of the 36 full-sib families and their parents. The pedigree simulation analysis by COLONY called for the average mutation rate, which we set as 5.0 x 103, based on the a previous publication on P. vannamei. Complete genotyping data for 16 loci in 1,152 shrimp samples were initially included for parentage analysis. Thereafter, numbers of loci were sequentially reduced in a randomized way for estimation of the minimum number of loci required for the various degrees of accuracy in parentage assignment, presumably with genotype error rate set at 0.01 in every run of the program [43-46].
Genetic variability and inbreeding
The statistical results for 36 families are summarized in, as all the genotypic data from 36 pairs of parents and their 1,080 individual offspring’s at 16 microsatellite loci were submitted for analysis. First, genetic variability of the parental and offspring groups were compared. NA in the parents’ group was 10.625, whereas that of the filial group was 10.052. The values of HO and HE among the parents were 0.891 and 0.804, respectively, and in the offspring, they were 0.813 and 0.792, respectively. Although their values were slightly higher in parents than in offspring, NA, HO, and HE revealed no significant difference between the parental and filial groups of all 36 families by FSTAT 2.9.3 (p>0.05). It is noteworthy that heterozygote deficiency was in negative value in each family, the average FIS of parents was slightly lower than that of offspring, from -0.107 to -0.026, but not significantly so. Second, the evaluation of genetic diversity in the offspring among the 36 families at 16 microsatellite loci was assessed with the same set of genetic parameters. The family NA values ranged from 2.50 to 3.25, HO from 0.710 and 0.915, and HE from 0.520 to 0.641.Within each family, the average inbreeding coefficient depended on specific locus, ranging 25 from -0.522 to -0.268. In the present study, no inbreeding occurred when viable families were generated from the selected brood stock (Tables 2-4).
| Genetic variation | |||||
|---|---|---|---|---|---|
| Group/Family | N | NA | HO | HE | FIS |
| Overall parents | 72 | 10.625 | 0.891 | 0.804 | -0.107 |
| Overall offspring | 1,080 | 10.052 | 0.813 | 0.792 | -0.026 |
| #1 | 30 | 2.875 | 0.817 | 0.576 | -0.443 |
| #2 | 30 | 2.625 | 0.777 | 0.561 | -0.398 |
| #3 | 30 | 3.063 | 0.821 | 0.602 | -0.373 |
| #4 | 30 | 2.75 | 0.763 | 0.567 | -0.341 |
| #5 | 30 | 2.75 | 0.754 | 0.545 | -0.366 |
| #6 | 30 | 2.688 | 0.852 | 0.581 | -0.492 |
| #7 | 30 | 3.25 | 0.873 | 0.632 | -0.387 |
| #8 | 30 | 2.875 | 0.856 | 0.6 | -0.443 |
| #9 | 30 | 2.938 | 0.915 | 0.617 | -0.505 |
| #10 | 30 | 3.063 | 0.867 | 0.612 | -0.433 |
| #11 | 30 | 2.75 | 0.825 | 0.565 | -0.459 |
| #12 | 30 | 2.5 | 0.765 | 0.527 | -0.474 |
| #13 | 30 | 2.75 | 0.84 | 0.585 | -0.451 |
| #14 | 30 | 2.75 | 0.835 | 0.557 | -0.522 |
| #15 | 30 | 2.875 | 0.879 | 0.59 | -0.502 |
| #16 | 30 | 2.813 | 0.808 | 0.552 | -0.451 |
| #17 | 30 | 2.938 | 0.819 | 0.577 | -0.422 |
| #18 | 30 | 2.75 | 0.783 | 0.559 | -0.402 |
| #19 | 30 | 2.813 | 0.763 | 0.567 | -0.35 |
| #20 | 30 | 2.875 | 0.804 | 0.557 | -0.456 |
| #21 | 30 | 2.813 | 0.779 | 0.547 | -0.422 |
| #22 | 30 | 3 | 0.833 | 0.606 | -0.372 |
| #23 | 30 | 2.875 | 0.792 | 0.561 | -0.409 |
| #24 | 30 | 2.75 | 0.792 | 0.561 | -0.418 |
| #25 | 30 | 2.938 | 0.879 | 0.605 | -0.473 |
| #26 | 30 | 3.063 | 0.833 | 0.594 | -0.398 |
| #27 | 30 | 2.938 | 0.821 | 0.568 | -0.438 |
| #28 | 30 | 2.688 | 0.731 | 0.562 | -0.288 |
| #29 | 30 | 3 | 0.815 | 0.581 | -0.407 |
| #30 | 30 | 3.125 | 0.91 | 0.641 | -0.436 |
| #31 | 30 | 2.938 | 0.804 | 0.583 | -0.371 |
| #32 | 30 | 3 | 0.869 | 0.594 | -0.485 |
| #33 | 30 | 2.875 | 0.833 | 0.603 | -0.396 |
| #34 | 30 | 2.813 | 0.719 | 0.559 | -0.269 |
| #35 | 30 | 2.875 | 0.71 | 0.52 | -0.339 |
| #36 | 30 | 2.75 | 0.723 | 0.567 | -0.268 |
Table 2: Comparison of genetic variability of 36 P. vannamei families at 16 microsatellites loci (N- sample size, Na- number of alleles, Ho- observed heterozygosity, HE- expected heterozygosity, FIS- inbreeding coefficient).
Family differentiation
The fixation index, FST, pairwise values among the 36 families, are presented in Table 3. The lowest FST was 0.157, between families #9 and #10, and the highest FST was 0.392, between families #19 and #5. The average of all pair wise FST values was 0.28. These values show that distinct genetic differentiation existed between any two families. In addition, the principal components analysis indicated that family differentiation could be divided between 9.93% for principal component 1 (PC1) and 8.12% for principal component 2 (PC2) among the offspring group. The additive genetic differentiation totaled at 18.05%. As shown in, the 36 families were distributed evenly and relatively randomly with respect to the two principal components’ axes, without noticeable familial cluster among the offspring. These results show that genetic differences of the families were well maintained, and inbreeding was under adequate control among all 36 offspring families when they were generated in this breeding program by following the mating design (Figure 1).
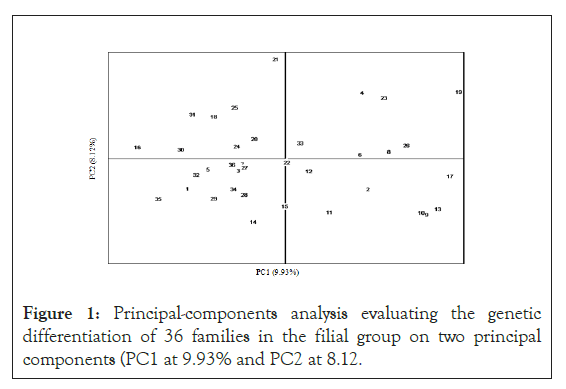
Figure 1: Principal-components analysis evaluating the genetic differentiation of 36 families in the filial group on two principal components (PC1 at 9.93% and PC2 at 8.12.
| Family | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 | #11 | #12 | #13 | #14 | #15 | #16 | #17 | #18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 #2 | 0.255 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| #3 | 0.213 | 0.268 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| #4 | 0.308 | 0.283 | 0.274 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| #5 | 0.256 | 0.287 | 0.222 | 0.296 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| #6 | 0.256 | 0.253 | 0.219 | 0.304 | 0.282 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| #7 | 0.192 | 0.244 | 0.183 | 0.235 | 0.229 | 0.253 | - | - | - | - | - | - | - | - | - | - | - | - |
| #8 | 0.272 | 0.278 | 0.219 | 0.285 | 0.295 | 0.201 | 0.234 | - | - | - | - | - | - | - | - | - | - | - |
| #9 | 0.271 | 0.210 | 0.265 | 0.282 | 0.294 | 0.283 | 0.220 | 0.256 | - | - | - | - | - | - | - | - | - | - |
| #10 | 0.274 | 0.222 | 0.258 | 0.280 | 0.280 | 0.213 | 0.226 | 0.229 | 0.157 | - | - | - | - | - | - | - | - | - |
| #11 | 0.271 | 0.274 | 0.190 | 0.327 | 0.301 | 0.271 | 0.216 | 0.204 | 0.225 | 0.200 | - | - | - | - | - | - | - | - |
| #12 | 0.329 | 0.331 | 0.277 | 0.316 | 0.297 | 0.295 | 0.252 | 0.253 | 0.265 | 0.285 | 0.257 | - | - | - | - | - | - | - |
| #13 | 0.278 | 0.239 | 0.274 | 0.285 | 0.257 | 0.250 | 0.248 | 0.236 | 0.201 | 0.161 | 0.229 | 0.282 | - | - | - | - | - | - |
| #14 | 0.212 | 0.239 | 0.238 | 0.342 | 0.317 | 0.301 | 0.239 | 0.263 | 0.243 | 0.237 | 0.233 | 0.302 | 0.284 | - | - | - | - | - |
| #15 | 0.271 | 0.279 | 0.217 | 0.345 | 0.303 | 0.260 | 0.271 | 0.246 | 0.266 | 0.255 | 0.248 | 0.323 | 0.252 | 0.254 | - | - | - | - |
| #16 | 0.235 | 0.331 | 0.240 | 0.338 | 0.312 | 0.315 | 0.213 | 0.301 | 0.326 | 0.303 | 0.271 | 0.298 | 0.338 | 0.261 | 0.304 | - | - | - |
| #17 | 0.346 | 0.268 | 0.292 | 0.304 | 0.331 | 0.277 | 0.299 | 0.281 | 0.223 | 0.207 | 0.275 | 0.284 | 0.200 | 0.306 | 0.281 | 0.338 | - | - |
| #18 | 0.277 | 0.336 | 0.271 | 0.300 | 0.283 | 0.264 | 0.277 | 0.317 | 0.323 | 0.298 | 0.340 | 0.331 | 0.307 | 0.311 | 0.290 | 0.264 | 0.299 | - |
| #19 | 0.338 | 0.309 | 0.294 | 0.291 | 0.392 | 0.255 | 0.296 | 0.223 | 0.283 | 0.288 | 0.317 | 0.341 | 0.300 | 0.337 | 0.337 | 0.340 | 0.276 | 0.324 |
| #20 | 0.282 | 0.290 | 0.239 | 0.288 | 0.232 | 0.255 | 0.212 | 0.266 | 0.286 | 0.292 | 0.306 | 0.275 | 0.295 | 0.315 | 0.325 | 0.283 | 0.327 | 0.282 |
| #21 | 0.323 | 0.340 | 0.308 | 0.312 | 0.360 | 0.311 | 0.311 | 0.319 | 0.352 | 0.327 | 0.335 | 0.366 | 0.362 | 0.356 | 0.338 | 0.321 | 0.319 | 0.277 |
| #22 | 0.297 | 0.299 | 0.262 | 0.304 | 0.290 | 0.297 | 0.253 | 0.234 | 0.268 | 0.284 | 0.280 | 0.262 | 0.277 | 0.279 | 0.251 | 0.314 | 0.290 | 0.283 |
| #23 | 0.339 | 0.297 | 0.311 | 0.297 | 0.343 | 0.305 | 0.306 | 0.230 | 0.303 | 0.296 | 0.297 | 0.360 | 0.313 | 0.322 | 0.291 | 0.334 | 0.312 | 0.328 |
| #24 | 0.290 | 0.288 | 0.260 | 0.295 | 0.255 | 0.293 | 0.228 | 0.299 | 0.274 | 0.291 | 0.291 | 0.291 | 0.298 | 0.296 | 0.309 | 0.305 | 0.324 | 0.307 |
| #25 | 0.290 | 0.290 | 0.257 | 0.281 | 0.277 | 0.311 | 0.221 | 0.295 | 0.306 | 0.298 | 0.287 | 0.312 | 0.302 | 0.327 | 0.283 | 0.270 | 0.300 | 0.290 |
| #26 | 0.273 | 0.267 | 0.279 | 0.274 | 0.327 | 0.264 | 0.252 | 0.232 | 0.239 | 0.239 | 0.254 | 0.308 | 0.259 | 0.318 | 0.326 | 0.336 | 0.294 | 0.329 |
| #27 | 0.261 | 0.289 | 0.246 | 0.310 | 0.271 | 0.269 | 0.246 | 0.255 | 0.278 | 0.242 | 0.258 | 0.304 | 0.294 | 0.255 | 0.259 | 0.257 | 0.274 | 0.266 |
| #28 | 0.236 | 0.285 | 0.255 | 0.322 | 0.328 | 0.268 | 0.213 | 0.268 | 0.294 | 0.258 | 0.232 | 0.326 | 0.299 | 0.279 | 0.263 | 0.281 | 0.305 | 0.319 |
| #29 | 0.236 | 0.262 | 0.265 | 0.356 | 0.302 | 0.287 | 0.267 | 0.304 | 0.265 | 0.271 | 0.287 | 0.341 | 0.318 | 0.266 | 0.258 | 0.287 | 0.286 | 0.265 |
| #30 | 0.252 | 0.265 | 0.243 | 0.286 | 0.230 | 0.263 | 0.186 | 0.269 | 0.278 | 0.268 | 0.256 | 0.233 | 0.288 | 0.254 | 0.257 | 0.198 | 0.267 | 0.258 |
| #31 | 0.237 | 0.281 | 0.241 | 0.269 | 0.264 | 0.286 | 0.229 | 0.242 | 0.313 | 0.300 | 0.291 | 0.309 | 0.322 | 0.271 | 0.306 | 0.236 | 0.350 | 0.268 |
| #32 | 0.238 | 0.303 | 0.243 | 0.311 | 0.269 | 0.280 | 0.244 | 0.277 | 0.296 | 0.268 | 0.252 | 0.272 | 0.280 | 0.272 | 0.305 | 0.265 | 0.335 | 0.258 |
| #33 | 0.265 | 0.271 | 0.234 | 0.291 | 0.330 | 0.231 | 0.244 | 0.221 | 0.278 | 0.255 | 0.262 | 0.297 | 0.283 | 0.273 | 0.288 | 0.282 | 0.306 | 0.312 |
| #34 | 0.229 | 0.276 | 0.241 | 0.312 | 0.294 | 0.263 | 0.257 | 0.252 | 0.265 | 0.255 | 0.251 | 0.299 | 0.267 | 0.235 | 0.266 | 0.275 | 0.286 | 0.269 |
| #35 | 0.256 | 0.312 | 0.261 | 0.369 | 0.294 | 0.315 | 0.277 | 0.304 | 0.304 | 0.279 | 0.272 | 0.324 | 0.306 | 0.259 | 0.246 | 0.277 | 0.340 | 0.310 |
| #36 | 0.292 | 0.306 | 0.229 | 0.301 | 0.293 | 0.318 | 0.244 | 0.265 | 0.278 | 0.285 | 0.251 | 0.321 | 0.306 | 0.273 | 0.248 | 0.271 | 0.311 | 0.327 |
Table 3: The pairwise FST values among 36 families of P. vannamei according to the genotypic data at 16 microseatellite loci.
Properties of microsatellite markers
To better elucidate the specific characteristics of these microsatellite loci, we included all 1,152 specimens’ genotyping data in the investigation. The results are summarized in Table 4. There were between 6 and 20 alleles per locus and 10.625 as average. Between 6 and 20 alleles were found for each locus, and 10.625 was the average. The minimum observed heterozygosity was 0.561, maximum 0.971, and overall mean 0.818. It was very close to the expected heterozygosity, the mean of 0.793, which valued between 0.596 and 0.926. Inbreeding coefficient ranged from -0.431 to 0.194, average -0.035. In terms of their exhibited polymorphism and heterozygosity, the selected microsatellite loci were fundamental for constructing the pedigrees of the 36 families. These values would also serve as useful baseline information for evaluating the SPF P. vannamei in successive generations in the breeding program.
| Locus | NA | HO | HE | FIS |
|---|---|---|---|---|
| TUGAPv 7-9.119 | 10 | 0.88 | 0.846 | -0.04 |
| TUMXLv 5.66 | 11 | 0.787 | 0.852 | 0.076 |
| TUMXLv 7.56 | 18 | 0.892 | 0.906 | 0.015 |
| TUMXLv 7.97 | 8 | 0.615 | 0.736 | 0.165 |
| TUMXLv 7.121 | 6 | 0.77 | 0.66 | -0.167 |
| TUMXLv 7.138 | 20 | 0.971 | 0.926 | -0.048 |
| TUMXLv 8.193 | 6 | 0.918 | 0.642 | -0.431 |
| TUMXLv 8.256 | 9 | 0.918 | 0.853 | -0.076 |
| TUMXLv 9.93 | 14 | 0.964 | 0.904 | -0.066 |
| TUMXLv 9.116 | 7 | 0.654 | 0.648 | -0.009 |
| TUMXLv 9.178 | 8 | 0.836 | 0.813 | -0.028 |
| TUMXLv 10.14 | 13 | 0.97 | 0.874 | -0.11 |
| TUMXLv 10.33 | 7 | 0.561 | 0.596 | 0.06 |
| TUMXLv 10.341 | 10 | 0.643 | 0.798 | 0.194 |
| TUMXLv 10.411 | 9 | 0.788 | 0.801 | 0.016 |
| TUMXLv 10.455 | 14 | 0.914 | 0.827 | -0.105 |
| Average | 10.625 | 0.818 | 0.793 | -0.035 |
Table 4: Summarized attributes of 16 microsatellite loci used in parentage analysis for two consecutive generations. Sample size was 1,154 for all. (NA- number of alleles, HO-observed heterozygosity, HE- expected heterozygosity, FIS- inbreeding coefficient).
Pedigree verification
The genotyping data of all 30 samples from each of the 36 families, together with those of their parents’, were used for parentage- assignment analyses. On the assumption that the genotyping error was 1%, the analyses for correct rate of parentage assignment by means of CERVUS 3.0 and COLONY 2.0 were conducted initially on the full data set, then with the data with 15 randomly selected loci, then from 14, then 13, etc., until data from only 6 loci remained in the final analysis. The marker-assisted pedigree was compared with the know pedigree for verification. The results from both programs indicated that the minimum loci needed for 100% success rate in assignment of an individual shrimp to its parents was 12. As the number of loci decreased from 12 to 6, accuracy declined. With 10 loci, it was 99.5% accuracy from either program, but with only 6, accuracy decreased to 80.65% in CERVUS and to 58.15% in COLONY. From these results, we conclude that any combination of 9 microsatellite loci from this panel would serve as reliable way to identify pedigree among the untagged P. vannamei stock on Guam (Figure 2).
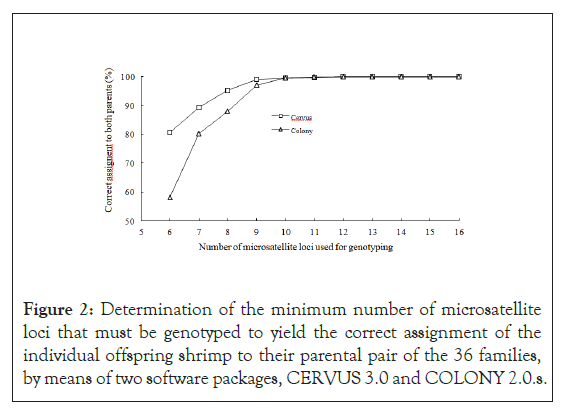
Figure 2: Determination of the minimum number of microsatellite loci that must be genotyped to yield the correct assignment of the individual offspring shrimp to their parental pair of the 36 families, by means of two software packages, CERVUS 3.0 and COLONY 2.0.s.
Good health status is commonly emphasized in most animal breeding programs because all the genetic improvement from generations of selection would be easily compromised or even totally lost if infectious disease broke out among the stock. Although it is not an inheritable trait, SPF health status with the genetically diverse founders is essential for a sustainable breeding program. Establishment of the SPF status requires a minimum of two years with strict health surveillance and bio security procedures. The shrimp stock from Guam’s hatchery started with SPF stocks as indicated by the USMSFP’s SPF stock protocols and has been monitored according to the updated lists annually for 12 years [13].
The SPF status of the shrimp stock in the breeding program has been successfully kept in the bio secure GADTC facility, but its genetic diversity had not been evaluated molecularly until the fourth and fifth generation’s shrimp families were included in our study. The panel of 16 highly polymorphic microsatellite loci selected from a pool of 128 loci has proved to be valuable in quantifying the genetic diversity, identifying the genetic differentiation among families, and verifying the pedigree of SPF P. vannamei. As was found in previous studies, properly selected microsatellite DNA markers were fundamental to evaluation of genetic variability, to genetic improvement strategies, and to pedigree construction in the white shrimp selective breeding programs. The average variation among 16 loci in our study was greater than those in previous studies of cultured populations of the same species, where different sets of microsatellite markers were used. In one study with 11 polymorphic microsatellite loci in domesticated P. vannamei, alleles per locus varied from 2 to 18, and observed heterozygosity ranged from 0.0286 to 0.9429, average 0.5610. In another study, using 8 loci, between 2 and 12 alleles at each locus, and heterozygosity from 0.20 to 0.73 were reported. In a third study that used 8 loci for monitoring of two generations which were different from the second study, between 7 and 10 alleles were recorded for each locus, and heterozygosity valued between 0.60 and 0.74, average 0.693. Decrease in genetic variation is demonstrated if the observed heterozygosity is significantly lower in the second generation than expected under Hardy-Weinberg equilibrium. In the present study, these two heterozygosities did not differ from one another. In contrast, when applying 4 microsatellite DNA loci for evaluating the genetic diversity, the properties of which were NA between 1 and 9 and Ne in the range of 1.1 and 6.72, Machado-Tamayo (2006) discovered loss of genetic variation because the observed heterozygosity ranged from 0.29 to 0.63 among the populations studied, and was distinctly lower than expected. The common cause of loss of genetic diversity is intensive selection within a small size of the broodstock pool for high performance on very few genetic traits of interest.In the present study, because the broodstock selection was processed under the guidance of the structured pedigree and genetic background, genotyping with the 16 highly polymorphic loci confirmed the high genetic diversity of the domesticated shrimp stock in the breeding program at GADTC [26,27,30,31,34,47].
Furthermore, we compared the genetic variation in our domesticated population with that of wild P. vannamei, as well as those of other penaeid species. A previous investigation of the genetic variation, which used 5 microsatellite DNA loci among the wild P. vannamei, collected at four different geographic locations in Mexico and Panama found from 2 to 16 alleles at each locus, 1.5 to 10.5 as the effective number of alleles, and observed heterozygosity ranging from 0.045 to 0.614, average of 0.320. The genetic variability in the wild stock was not found to be higher than that of the domesticated stock from the present study, but because completely different panels of microsatellite loci were used for in the two studies in P. vannamei, comparison of the two is difficult [48].
More alleles were reported in wild tiger shrimp from Australia: 34 to 84 per locus but from only three loci. Ten loci were employed in another study to investigate eight wild tiger shrimp populations in India, and results revealed 6 to 49 alleles at each locus, and 0.0750 to 1.0 for the observed heterozygosity. Similarly, in four wild populations of tiger shrimp in the Philippines, six loci showed 6 to 54 alleles each locus, and heterozygosity ranged from 0.47 to 1.0. The much higher allelic number and heterozygosity were found in wildstock of P. monodon (tiger shrimp) than in P. vannamei. Used 5 microsatellite loci to analyze five wild populations of P. monodon from Thailand, and reported 19 to 30 alleles at each locus, and heterozygosity from 0.49 to 0.95. More alleles were documented in wild tiger shrimp from Australia: 34 to 84 per locus but from only three loci. Ten loci were employed in another study to investigate eight wild tiger shrimp populations in India, and results revealed 6 to 49 alleles at each locus, and 0.0750 to 1.0 for the observed heterozygosity. Similarly, in four wild tiger shrimp populations in the Philippines, six loci showed 6 to 54 alleles each locus, and heterozygosity ranged from 0.47 to 1.0. The parameters of genetic variance were broader in P. monodon than in P. vannamei in general. Contributing factors could be the genetic makeup of the species, the scope of the population being investigated, and the characteristics of the species-specific loci being selected [49-51].
A sustainable breeding program needs to balance genetic gains through selection, in the genetic trait of current interest and the maintenance of adequate genetic diversity to permit selection for other heritable traits in the future. The negative impacts of inbreeding must therefore be minimized, but substantial genetic gain can be achieved over generations of selection in the meantime without abrupt loss of genetic diversity within the domesticated population. Furthermore, genetic variation is related to fitness and adaptability of a cultured population, so proper evaluation is important because genetic diversity among the selected broodstock would directly lead to the sustainable production of high-quality seed stock. Consequently, the continual monitoring of genetic variation in each consecutive generation, by means of molecular markers, is a feasible breeding-program practice that permits quantifying the changes in variability caused by inbreeding, high intensity of selection, genetic drift, bottleneck effects, etc [52-59].
The loss of genetic variation over multiple generations in the domesticated population has been clearly documented for various penaeid species. Comparison of data from 20 allozyme loci for six consecutive generations of P. japonicus revealed that heterozygosity in the population was progressively reduced from 0.102 to 0.039, the lowest effective number of parents for generating the broodstock was four individuals. Breeding among a small number of the parents and execution of inadequate mating strategies are the common causes of the loss of genetic variability in cultured stocks and are associated with genetic drift or inbreeding. Similarly, the loss of genetic diversity due to reduced effective population size has also been reported in P. vannamei, P. stylirostris, and P. monodon. Because small effective population sizes and poor management were the two primary reasons for the loss of genetic diversity, more research is needed to establish the population size sufficient to reduce inbreeding and keep the genetic diversity at acceptable levels. Allendorf and Ryman have suggested several considerations for establishing the broodstock in a way that avoids significant depletion of genetic variability. These principles were applied in the breeding program at GADTC, which started with a was sizeable and genetically diversified founder stock, prompting a higher effective breeding number by using equal proportions of male and female breeders, and mating the unrelated individuals by pedigree. Analysis based on genotyping with the microsatellite loci showed that the average inbreeding coefficients (FIS) of parent families and offspring families were -0.107 and -0.026, respectively, verifying the absence of inbreeding in either generation. Although the panel of microsatellite loci proves to be a useful tool for maintaining the genetic diversity of shrimp stock at GADTC, several other methods using molecular markers for genetic analysis had also been scrutinized according to previously published results. Zhen suggested that selective breeding in shrimp stocks could deviate from the Hardy-Weinberg equilibrium. Stated that the heterozygosity deficit could result from the presence of null alleles. Error in allelic scoring due to stutter bands has been reported for L. setiferus and P. monodon. These technical artifacts could misclassify heterozygotes as homozygotes because the alleles are so close in size. Only highly polymorphic microsatellite loci with distinctly strong bands were chosen for the panel and scored in our study, so the adverse effects caused by the stutter bands and null alleles were eliminated. In addition, the estimated values of FST ranging from 0.157 to 0.392 among 36 offspring families indicated that the offspring groups differed in genetic relationships, and the genetic structure among families might differ as well. In selective breeding programs of aquatic species with high fecundity, like shrimp, a detailed record of pedigree information over multiple generations is critical. Color-coded elastomer implants were used to tag individuals and maintaining familial identification in our study, as well as in several others [37,51,56,60-69].
These crustaceans had needed to be cultivated separately by the family until they were big enough for addition of elastomer tags. The drawbacks of this tagging method are that it is time consuming and labor intensive and is restricted by availability of holding facilities, concern for environmental interactions, and the size and number of animals to be tagged. Because it can be applied noninvasively and after the phenotypic data are collected at the end of the mixed-culture period, DNA fingerprinting for parentage analysis could overcome the constrains imposed by physical tagging. Microsatellite loci offer a good alternative for maintaining the genealogical linkage, and could potentially replace the traditional method for family identification in aquaculture breeding programs. Microsatellite DNA markers are suitable for pedigree identification because they are well known for their codominance, Mendelian inheritance, abundant presence in the genome, and high levels of polymorphism. Although several limiting factors could interfere with their power and accuracy in the parentage assignment-mutation, null alleles, and genotyping errors microsatellite technique remained as a useful tool for studying parentage in some aquatic species. In the study reported here, any combination of 12 and more microsatellite loci perfectly matched 1,080 individual shrimp with their 36 sets of parents, and 10 loci produced over 99.5% accuracy. The success rate and sample size of our parentage analysis was much higher than those previously reported in two other penaeid shrimp. Rrelied on five microsatellite loci to analyze the parentage in Fenneropenaeus chinensis and achieved only 92.9% accuracy of assignment of progeny to their real parental couple, and in a parentage study P. japonicus, only 47% of progeny were assigned correctly to their dams according to six microsatellite loci’s genotyping data. The use of a sufficient number of polymorphic microsatellite loci would boost the accuracy in parentage identification. In Macrobrachium rosenbergii (freshwater prawn), success rates of 95.6% and 95.2% in pedigree identification were achieved with 9 microsatellite markers by COLONY and CERVUS, respectively. Use of eight informative microsatellite loci yielded an accuracy of 98% in linking individual Salmo salar (Altantic salmon) of the 10 full-sib families with their parental pairs, whereas 15 loci yielded 100% accuracy. In a study with Cyprinus carpio L. (common carp), only microsatellite markers yield an accuracy of 95% for offspring from 240 crosses. Use of genetic profiling to identify parentage and/or genetic relatedness would reinforce the minimization of potential adverse effects of accumulated inbreeding by allowing close monitoring of changes in genetic variation each generation, even while selection was used to maximize desirable phenotypic traits in the aquaculture breeding [20,45,70-78].
Both genetic diversity and pedigree assessments must be closely monitored and well maintained in shrimp breeding programs. Our panel comprising 16 microsatellite DNA markers in this study demonstrated its effectiveness in assessing genetic diversity and high efficacy of parentage assignment as it served in the SPF P. vannamei breeding program in Guam. Among 36 mated pairs and their offspring, the application of this microsatellite panel was useful for guiding the critical decision-making process during the selection and improvement of the stock utilization or management in the shrimp breeding program. Our findings have established a foundation for further development and implementation of more suitable, and ready to use, genetic markers in the continuous selective breeding program.
Grant support for this project was from USDA/NIFA program No. 2009-34135-2009 and USDA HATCH Project. No. GUA0559.The Guam Aquaculture Development and Training Center from the Western Pacific Tropical Research Center at the University of Guam provided the experimental shrimp, support of the shrimp breeding platform, and research facilities for this study. Kasetsart University granted the Student Exchange fellowship to Miss Channarong that permitted her to participate in research at University of Guam. We would like to thank Dr. James Grasela and Ms. A.B. Thistle for editing this paper.
The authors declared no conflict of interest.
Citation: Jiang HG, Channarong J, Ngernsiri L, Swatdipong A (2021) Microsatellite Techniques in Guam’s Specific-Pathogen-Free Penaeus Vannamei Stock: Genetic Variance and Parentage Identification. Fish Aquac J. S2: 003.
Received: 21-May-2021 Accepted: 04-Jun-2021 Published: 11-Jun-2021 , DOI: 10.35248/2150-3508.21.s2.003
Copyright: %copy; 2021 Jiang HG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.