Mycobacterial Diseases
Open Access
ISSN: 2161-1068
ISSN: 2161-1068
Research Article - (2023)Volume 13, Issue 1
Background: Mycobacteria could invade the host’s immune system to survive and persist in the host through different mechanisms such as the expression modulation of microRNAs (miRNAs). microRNA is a small, noncoding oligonucleotide that regulates gene expression and transcription. miRNAs' differential expressions in disease phenomena can serve as biomarkers. The expression level of serum-derived exosomal miRNAs from mycobacteria patients could result to enhance monocyte cell apoptosis. This study tries to evaluate four serumderived exosomal miRNAs as a potential mycobacterial biomarker.
Methods: Serum-derived exosomes were purified from serum samples of 55 patients with pulmonary tuberculosis or non-tuberculosis and 30 healthy controls. The expression level of Hsa-miR-20a-5p, Hsa-miR-29a, Hsa-miRlet7e, and Hsa-miR-155 was evaluated using qRT-PCR.
Results: In TB patients, the expression level of miR-20a-5p, miR-29a, and miR-let7e were significantly increased (p ≤ 0.0001), but the miR-155 expression level decreased. The receiver operating characteristic (ROC) curve analysis determined effective diagnostic biomarkers of miRNAs with an AUC=0.6933 for miR-20 (p ≤ 0.01), AUC=0.6011 for miR-29a (p ≤ 0.17), AUC=0.7322 for miR-let7e: (p ≤ 0.002), and AUC=0.7456 for miR- 155 (p ≤ 0.001) for active tuberculosis. The expression of miR-let7e, 20a, and 29a in Mycobacterium avium vs. Mycobacterium tuberculosis was up regulated (p ≤ 0.01, p ≤ 0.0001, and P ≤ 0.0001, respectively), same as miRs let7e and 20a expression which was increased in Mycobacterium abscessus vs. Mycobacterium tuberculosis (P ≤ 0.0001 and P ≤ 0.002, respectively).
Conclusion: In conclusion, circulating exosomal microRNA miR-20, miR-let7e, and miR-155 have diagnostic potential for active pulmonary tuberculosis. Furthermore, the study facilitates the development of potential biomarkers of pulmonary tuberculosis and non-tuberculosis.
Mycobacterium tuberculosis; Non-tuberculous mycobacteria; Exosomes; Apoptosis; microRNAs
Mycobacterium tuberculosis (Mtb) and Non-Tuberculous Mycobacteria (NTM) are still the most leading infection cause of global health problems and death. Despite global advances in health and medicine, tuberculosis remains an important global health challenge [1]. Based on WHO reports, a total of roughly 1.4 million deaths were reported from TB in 2019 and 7.1 million people were newly diagnosed with TB in 2019 [2]. Based on the report of tracking mycobacteria species by Mycobacterium Jones in 2018, Mycobacterium abscessus, Mycobacterium kansasi, Mycobacterium chelonae, Mycobacterium simiae, and Mycobacterium avium complex were most frequently encountered in clinical samples [3, 4] because of the restrictions in TB diagnostic methods and the lack of a robust method to differentiate with non-tuberculosis mycobacteria, clinicians are still faced with the challenge of early diagnosis. Early detection markers of Mycobacterium tuberculosis (Mtb) and differentiation other than Mycobacterium tuberculosis (Mtb) play an important role in controlling the disease and preventing infections transmission, so introducing any effective biomarkers will be extremely valuable [5]. Exosomes are novel diagnostic biomarkers used in different diseases such as cancers and infectious diseases [6, 7]. They are 30-150 nm vesicles released from different cell types and can be found in most human biofluids [8-10]. Exosomes have a role in cell-to-cell communication as they transfer biological information in the form of different molecules including microRNAs (miRNAs) between cells [11, 12]. MiRNAs are small 18-22 nucleotide non-coding RNAs that negatively regulate gene expression post-transcriptionally. microRNAs play a pivotal role in the most critical biological functions, and their dysregulation can lead to several pathological outcomes [13]. A systemically reviewed by Laura Moody showed quantification of miR-20a-5p as a Colorectal Cancer (CRC) biomarker. They reviewed 32 studies with a total of 5014 tumor cases and 2863 healthy subjects that expression level of miR-20a-5p was quantified in feces, serum, or tumor tissue. They found that miR-20a-5p was up-regulated in 20 studies of CRC patients versus controls. A pooled Area Under the Receiver Operating Characteristic Curve (AUROC) was computed to predict the sensitivity and specificity of miR-20a. miR-20a-5p might consider as a relevant biomarker for the detection of CRC [14].
In addition, miRNAs are associated with regulating inflammatory courses during bacterial infections like mycobacteria. Mycobacterium tuberculosis infection associate with host physiological responses including host immune and metabolic re-patterning [15] that maintain Mycobacterium tuberculosis (Mtb) to their nutritional needs and energy requirements, also increase their intracellular survival [16]. This involves the modulation of host miRNAs that controls the regulatory networks which associate with carbon, nitrogen, and lipid metabolism of the infected cells [17]. Some studies have reported a change in the expression signature of circulating and cellular miRNAs in patients with TB compared to healthy controls [18]. These mechanisms could be including induction of apoptotic pathway and autophagy, stimulation of IFN-γ, and Tumor Necrosis Factor Alpha (TNF-α) secretion which are adopted by the host cells during mycobacterial infection [19]. As an example, triggering cell apoptosis is a defense mechanism against intracellular pathogens, which are regulated by Mycobacterium tuberculosis modulated host miRNAs, Hsa-miR-29a, Hsa-miR-let7e, Hsa-miR-155, and Hsa- miR-20a-5p [20].
miR-29a was determined by different experiments as highly up regulated in patients with active TB vs. healthy controls and considering as a relevant miRNA in TB pathogenesis since directly or indirectly target IFN-γ production. So, miR-29a suppresses the host's innate and adaptive immune response against intracellular pathogens. There is some evidence that let-7 has down regulated in Mycobacterium tuberculosis infected macrophages at thetranscriptional level by Mycobacterium tuberculosis secreted 6 kilodalton (kDa) early secretory antigenic target (ESAT-6) that is a modulator of host immune responses and promote Mycobacterium tuberculosis escape from the phagosome [21,22]. miR-155 is dysregulated in human macrophages infected by different Mycobacterium tuberculosis and results to induce cell proliferation by inhibiting apoptosis. In Mycobacterium tuberculosis, innate immune cell activation is regulated by miR-155 and inhibits apoptosis in monocytes. Zhang et al. reported that there was a negative association between serum miR-155 abundance and NK cell cytotoxicity [23]. Serum miR-155 levels were in negative relation with the TB-suppressing activity of NK cells. These data support the importance of a single miRNA to target multiple genes at the same time and emphasizing the continuous study on the regulatory role of miRNAs [24]. Down regulating miR-20a in THP-1 macrophages induces apoptosis and increases the survival of Mtb [23]. Overexpression of miR-29a was confirmed as a candidate biomarker to discriminate between active TB and latent TB infection in a study from Cameroon [25]. So, Mycobacterium tuberculosis infection accelerates a dysregulation in the expression of those miRNAs involved in several physiological pathways of immunity, inflammation, and particularly cellular apoptosis which could be used as diagnostic and prognostic biomarkers and responses to therapy [20]. , Here we evaluate the power of serum derived-exosomal miRNAs of Hsa-miR-29a, Hsa-miR-let7e, Hsa-miR-155, and Hsa-miR-20a-5p as a potential biomarker in Mycobacterium tuberculosis and NTM infection.
Patient samples
In the current study, we evaluated 55 hospitalized patients who were diagnosed with mycobacterial infections including Mycobacterium abscessus, Mycobacterium kansasi, Mycobacterium chelonae, Mycobacterium simiae, Mycobacterium avium, and Mycobacterium tuberculosis. These individuals aged from 32 to 83 years olds were admitted to the Masih Daneshvari Hospital, a referral center for tuberculosis and lung diseases in Tehran-Iran, during 2018-2020. The inclusions criteria were the new cases of TB with positive Mtb culture or Mtb smear results. For NTM patients, the inclusions criteria were the clinical and radiological findings corresponding with NTM which were confirmed with appropriate molecular diagnosis or culture according to the official ATS/ IDSA statement [26]. The patients infected with Hepatitis B Virus (HBV), Hepatitis C Virus (HCV), or Human Immunodeficiency Virus (HIV) and those with a history of diabetes, haemoptysis, and smoking were excluded. Also, 30 healthy control individuals aged from 30 to 80 were studied for exosomal miRNA analysis and apoptosis induction. These healthy controls didn’t have any history of infection and chronic disease; they were evaluated for TB, HIV, and chronic disease by CRP and ESR assays, considering review past medical history. An institutional ethical board of Masih Daneshvari Hospital approved the current study (SBMU 1.REC1394.137).
Isolation and characterization of serum-derived exosomes
Exosomes were extracted from serum samples of patients infected with mycobacterial species and healthy subjects using exoEasy Maxi Kit (Qiagen, Valencia, CA, USA, Cat. no. 76064) according to the manufacturer’s instructions. Only 1 volume (1000 µl) of buffer XBP was added to 1 volume (1000 µl) of the sample, mixed by inverting, and left the mixture at room temperature. The mixture was then transferred to the exoEasy spin column and centrifuged at 500 × g for 1 min followed by discarding flow-through and placed the column into the collection tube. Next, 10 ml buffer XWP was added to the column and centrifuged at 3000 × g. Flow-through was discarded, 800 µl of buffer XE was added, incubated for 1 min, centrifuged at 500 × g for 5 min, and finally, the flow-through was collected as exosome. Purified exosomes were stored at -70 °C for size and morphology analysis by Transmission Electron Microscopy (TEM) (Zeiss-EM10C-100 KV, Germany), and Dynamic Light Scattering (DLS).
Exosomal RNA extraction and cDNA synthesis
Total RNA was isolated of the exosomes released from the serum of the mycobacterial patients using the Favor Prep miRNA kit according to the manufacturer’s instructions. First, 200 µl Lysis Buffer was added into the tube containing 1 × 10 cultured cell pellet followed by vortexing and incubating at room temperature for 10 minutes. Next, 20 µl 2M NaOAc, pH 5.2 was added and then inoculated with 180 µl ddH2O saturated phenol and also 40 µl chloroform, mixed vigorously for 2 minutes. The mixture was centrifuged at 3000 × g or 3 minutes and the upper phase was transferred into a clean tube. Ethanol to 35% volume was mixed well. The mixture was transferred to the RNA column in the collection tube and incubated for 1 minute and then centrifuged at 3000 × g for 30 seconds and the filtrate was collected. Ethanol to 70% volume was added and mixed properly. The mixture was then transferred to another RNA column in the collection tube and incubated for 1 minute followed by centrifuging at 3000 × g for 30 seconds. miRNA was bound to the column membrane and washed with 200 µl wash buffer 2 and incubated for 1 minute. To remove the residue liquid, another centrifuging at 3000 × g for 1 minute was performed. RNA column to a clean 1.5 ml tube was replaced and 50 µl release buffer was added and then incubated for 3 minutes. It was then centrifuged at 3000 × g for 3 minutes to obtain miRNA. The purified miRNA was kept at -70 °C for further analysis. Only 20 ng of the total RNA was reverse transcribed using the Yekta Tajhiz cDNA Synthesis Kit (Cat No: YT4500, Iran) according to the manufacturer’s instructions. Loop primers for each miRNAs candidate and a one-step of thermal cycler includes at 42 °C for 60 minutes, at 70 °C for 5 minutes were performed to complete cDNA synthesis.
Real-time PCR and miRNA quantification
Real-time PCR was performed using 2x qPCRBIO SYBR® Green Master Mix kit (Biosystems) for Hsa-miR-29a, Hsa-miR-let7e, Hsa- miR-155, and Hsa-miR-20a-5p according to the manufacturer’s instructions. Universal primer and specifically designed loop RT primers were used in all experiments. The cDNA product was diluted 10x and added to the real-time PCR reactions. A two-step real-time PCR protocol was done using an initial denaturation step at 95Ë?C for 2 min, a total of 50 amplification cycles including a denaturation step at 95 °C for 10 s, and an annealing step at 30Ë?C for 63 s. A melting curve was found for each reaction to approve the accuracy of the reactions. Expression levels of all the miRNAs were normalized according to the expression level of U6 as an endogenous control using the 2-â??â??Ct method.
Statistical analysis
All experiments were statistically analyzed for significant difference (p<0.05) using analysis of one-way variance ANOVA. The difference in gene expression was calculated using Genex-6 software and the graph was plotted using graph pad prism (v.8). The association between serum derived-exosomal miRNAs and increased serum apoptosis was evaluated using the sensitivity, specificity, and area under the Receiver Operating Characteristic (ROC) curve. The Area Under the Curve (AUC) was resolved with a 95% Confidence Interval (CI) and ROC analysis was performed using SPSS (v.21). Relative expression levels were calculated via the 2-â??â??Ct method.
Characterization of serum-derived exosomal miRNAs
Exosomes were extracted from the serum of the patients and healthy control subjects; morphologically confirmed by Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS) (Figures 1A and 1B). These exosomes expressed high levels of the CD81 exosomal marker protein, as determined by flow cytometry (Figure 1C).
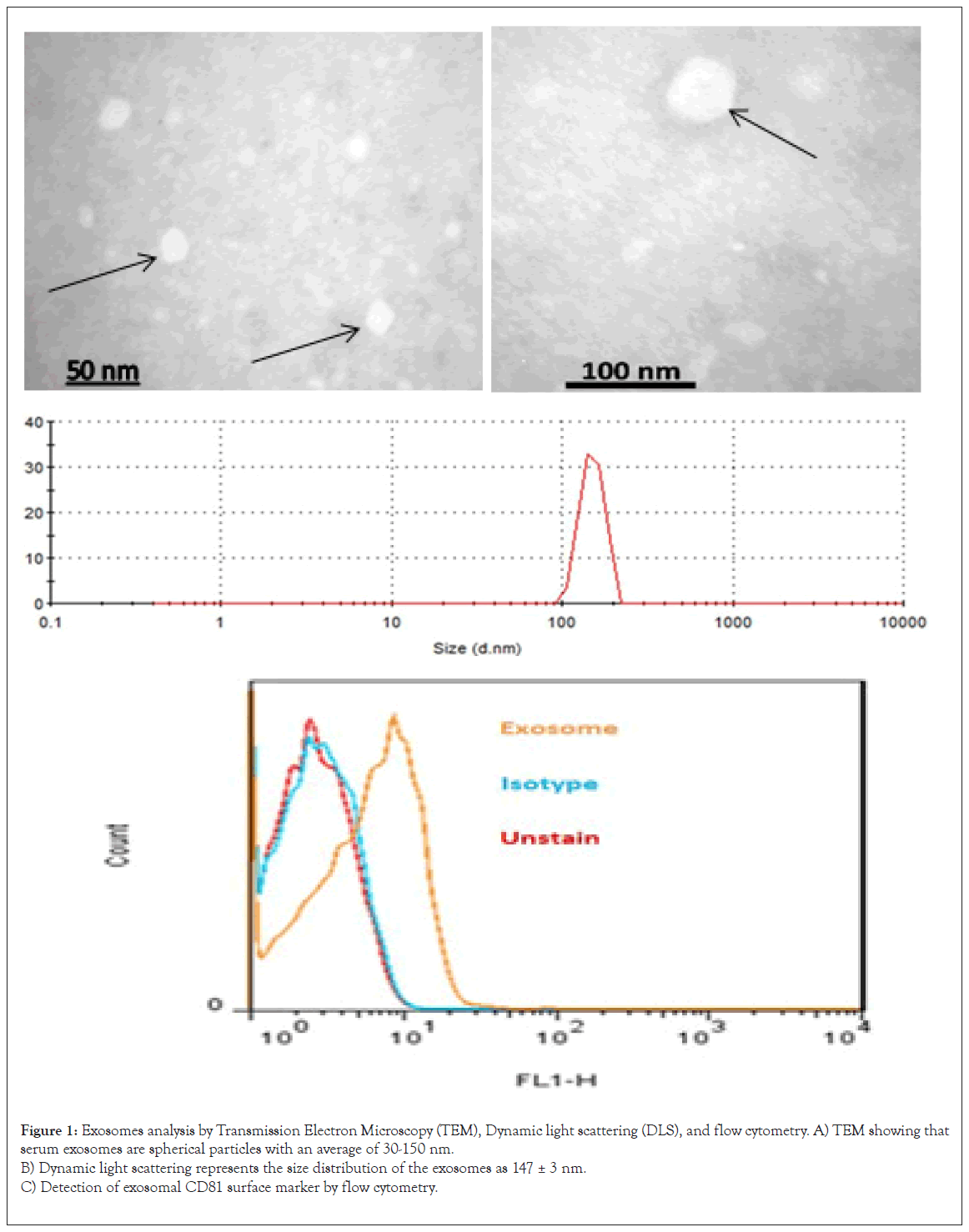
Figure 1: Exosomes analysis by Transmission Electron Microscopy (TEM), Dynamic light scattering (DLS), and flow cytometry.
A) TEM showing that
serum exosomes are spherical particles with an average of 30-150 nm.
B) Dynamic light scattering represents the size distribution of the exosomes as 147 ± 3 nm.
C) Detection of exosomal CD81 surface marker by flow cytometry.
Relative expression of Hsa-miR-29a, Hsa-miR-let7e, Hsa- miR-155 and Hsa-miR-20a-5p in both MTB and NTM compared to healthy controls
The relative expression of nuclear RNA U6 as an endogenous control was used for all experiments. After normalization with U6, up regulation of miR-20a-5p was statistically significant in Mycobacterium tuberculosis, Mycobacterium avium, Mycobacterium simiae, Mycobacterium chelonae, Mycobacterium abscessus respectively, vs. healthy controls (p ≤ 0.03, p ≤ 0.003, p ≤ 0.7, p ≤ 0.03, p ≤ 0.01 and p ≤ 0.008, respectively), not significantly for Mycobacterium kansasi (p ≤ 0.7). For Hsa-miR-29a, up regulations were significantly detected in Mycobacterium avium (p ≤ 0.01), Mycobacterium kansasi (p ≤ 0.01), and Mycobacterium chelonae (p ≤ 0.002), while it was not significantly in Mycobacterium tuberculosis (p ≤ 0.3) and Mycobacterium simiae (p ≤ 0.7) vs. healthy controls. A similar significant up regulation expression of miR-let7e was also detected in Mycobacterium tuberculosis (p ≤ 0.01), Mycobacterium avium (p ≤ 0.005), Mycobacterium kansasi (p ≤ 0.01), and Mycobacterium simiae (p ≤ 0.01), no in Mycobacterium chelonae (p ≤ 0.21). In contrast, only the expression level of Hsa-miR-155 was down-regulated. Statistically significant downregulation of Hsa- miR-155 was found in Mycobacterium tuberculosis (p ≤ 0.006) and Mycobacterium kansasi (p ≤ 0.03), but not in Mycobacterium avium (p ≤ 0.9), Mycobacterium simiae (p ≤ 0.9), Mycobacterium chelonae (p ≤ 0.9), and Mycobacterium abscessus (p ≤ 0.7) healthy subjects. The expressions of the miRNAs are summarized in Table 1 and Figure 2.
| miRNAs | Groups | Mean | P.value | fold | Description |
|---|---|---|---|---|---|
| miR-20a | M. tuberculosis vs. Control subjects | 1.38518 | 0.03031 | 2.61205 | Upregulated:2.612fold |
| M. avium vs. Control subjects | 3.12999 | 0.00394 | 8.75429 | Upregulated:8.7543fold | |
| M. kansasi vs. Control subjects | 1.26326 | 0.70434 | 2.40038 | Upregulated:2.4004fold | |
| M. simiae vs. Control subjects | 2.56852 | 0.0328 | 5.93201 | Upregulated:5.932fold | |
| M. chelonae vs. Control subjects | 2.88251 | 0.01049 | 7.37432 | Upregulated:7.3743fold | |
| M. abscessus vs. Control subjects | 2.94867 | 0.00813 | 7.72037 | Upregulated:7.7204fold | |
| M. avium vs. M. tuberculosis | 1.74482 | 0.32792 | 3.35153 | Upregulated:3.3515fold | |
| M. kansasi vs. M. tuberculosis | -0.12192 | 1.00000 | 0.91896 | Down regulated:-1.0882fold | |
| M. simiae vs. M. tuberculosis | 1.18335 | 0.76351 | 2.27104 | Upregulated:2.271fold | |
| M. chelonae vs. M. tuberculosis | 1.49734 | 0.51555 | 2.82322 | Upregulated:2.8232fold | |
| M. abscessus vs. M. tuberculosis | 1.56349 | 0.46241 | 2.95568 | Upregulated:2.9557fold | |
| miR-29a | M. tuberculosis vs. Control subjects | 0.9529 | 0.30207 | 1.93576 | Upregulated:1.9358fold |
| M. avium vs. Control subjects | 2.76987 | 0.01593 | 6.82046 | Upregulated:6.8205fold | |
| M. kansasi vs. Control subjects | 2.82456 | 0.01299 | 7.08398 | Upregulated:7.084fold | |
| M. simiae vs. Control subjects | 1.19216 | 0.75677 | 2.28495 | Upregulated:2.2849fold | |
| M. chelonae vs. Control subjects | 3.23078 | 0.00258 | 9.38775 | Upregulated:9.3878fold | |
| M. abscessus vs. Control subjects | 1.27237 | 0.69679 | 2.41558 | Upregulated:2.4156fold | |
| M. avium vs. M. tuberculosis | 1.81698 | 0.28028 | 3.52343 | Upregulated:3.5234fold | |
| M. kansasi vs. M. tuberculosis | 1.87166 | 0.24758 | 3.65953 | Upregulated:3.6595fold | |
| M. simiae vs. M. tuberculosis | 0.23926 | 0.99994 | 1.18039 | Upregulated:1.1804fold | |
| M. chelonae vs. M. tuberculosis | 2.27788 | 0.08343 | 4.84965 | Upregulated:4.8496fold | |
| M. abscessus vs. M. tuberculosis | 0.31947 | 0.99968 | 1.24787 | Upregulated:1.2479fold | |
| miR-let7e | M. tuberculosis vs. Control subjects | 1.23888 | 0.01276 | 2.36015 | Upregulated:2.3602fold |
| M. avium vs. Control subjects | 2.50717 | 0.00519 | 5.68504 | Upregulated:5.685fold | |
| M. kansasi vs. Control subjects | 2.2793 | 0.0152 | 4.85442 | Upregulated:4.8544fold | |
| M. simiae vs. Control subjects | 2.33794 | 0.01162 | 5.0558 | Upregulated:5.0558fold | |
| M. chelonae vs. Control subjects | 1.57966 | 0.21638 | 2.98899 | Upregulated:2.989fold | |
| M. abscessus vs. Control subjects | 3.18599 | 0.00014 | 9.10078 | Upregulated:9.1008fold | |
| M. avium vs. M. tuberculosis | 1.26829 | 0.47382 | 2.40876 | Upregulated:2.4088fold | |
| M. kansasi vs. M. tuberculosis | 1.04042 | 0.69852 | 2.05683 | Upregulated:2.0568fold | |
| M. simiae vs. M. tuberculosis | 1.09906 | 0.64192 | 2.14215 | Upregulated:2.1422fold | |
| M. chelonae vs. M. tuberculosis | 0.34079 | 0.99856 | 1.26645 | Upregulated:1.2664fold | |
| M. abscessus vs. M. tuberculosis | 1.94712 | 0.06142 | 3.85604 | Upregulated:3.856fold | |
| miR-155 | M. tuberculosis vs. Control subjects | -1.03761 | 0.00676 | 0.48713 | Down regulated:-2.0528fold |
| M. avium vs. Control subjects | -0.57946 | 0.92378 | 0.66921 | Down regulated:-1.4943fold | |
| M. kansasi vs. Control subjects | -1.655 | 0.03445 | 0.31754 | Down regulated:-3.1492fold | |
| M. simiae vs. Control subjects | -0.59859 | 0.91198 | 0.6604 | Down regulated:-1.5142fold | |
| M. chelonae vs. Control subjects | -0.36356 | 0.99251 | 0.77724 | Down regulated:-1.2866fold | |
| M. abscessus vs. Control subjects | -0.75147 | 0.78001 | 0.594 | Down regulated:-1.6835fold | |
| M. avium vs. M. tuberculosis | 0.45815 | 0.97509 | 1.37378 | Upregulated:1.3738fold | |
| M. kansasi vs. M. tuberculosis | -0.61739 | 0.89933 | 0.65185 | Down regulated:-1.5341fold | |
| M. simiae vs. M. tuberculosis | 0.43903 | 0.97991 | 1.35569 | Upregulated:1.3557fold | |
| M. chelonae vs. M. tuberculosis | 0.67405 | 0.85491 | 1.59555 | Upregulated:1.5955fold | |
| M. abscessus vs. M. tuberculosis | 0.28615 | 0.99799 | 1.21938 | Upregulated:1.2194fold |
Table 1: Expression level changes of serum exosomal miRNAs Has-miR-20a-5p, Has-miR-29a, Has-miR-let7e, and Has-miR-155 individually and in a combination of each mycobacterial species.
Figure 2: The relative expression of exosomal miR-20a-5p, miR-29a, miR-let7e, and miR-155 in Mycobacterium tuberculosis and non-tuberculous mycobacteria patients compared to healthy controls. Real-time PCR of exosomal miR-20a-5p, miR-29a, miR-let7e indicated up regulation in the patients compared to healthy controls (*p<0.05, **p<0.001, and ***p<0.0001 respectively). But, miR-155 determined down regulation in comparison with healthy controls. Up regulation of exosomal miR-20a-5p, miR-29a, miR-let7e was found, however, miR-155 down regulated. Data represent mean ± SEM from 55 mycobacteria patients and 30 control subjects. Note:  Non-tuberculous mycobacteria
Non-tuberculous mycobacteria
qRT-PCR of Hsa-miR-29a, Hsa-miR-let7e, Hsa-miR-155 and Hsa-miR-20a-5p in NTM compared to MTB
The expression level of the miRNAs in NTM species was compared to Mtb to evaluate as a potential biomarker. The expression of Hsa-miR-20a-5p, Hsa-miR-29a, and Hsa-miR-let7e was statistically significant in Mtb patients (p ≤ 0.0001, P ≤ 0.0001, P<0.0001, respectively) (Table 2 and Figure 3). No statistical significance was found in the Hsa-miR-155 expression of NTM vs. Mtb P ≤ 0.2). Based on Tukey test analysis, significant statistics analysis was found for Hsa-miR-let7 in Mycobacterium avium vs. Mtb (p ≤ 0.01). The other statistical results of the evaluated miRNAs are summarized in Table 1.
| Groups | miRNAs | Fold change | SD | Lower 95% CI of mean | Upper 95% CI of mean |
|---|---|---|---|---|---|
| M. tuberculosis (n=30) | let.7 | 1.000000 | 1.779672 | 0.6061 | 1.394 |
| M. avium (n=5) | let.7 | 2.408758 | 0.27311 | 1.597 | 3.22 |
| M. kansasi (n=5) | let.7 | 2.05683 | 0.375233 | 1.597 | 2.517 |
| M. simiae (n=5) | let.7 | 2.142151 | 0.363644 | 1.695 | 2.589 |
| M. chelonae (n=5) | let.7 | 1.266447 | 0.27001 | 0.9367 | 1.596 |
| M. abscessus (n=5) | let.7 | 3.856029 | 0.70682 | 2.99 | 4.722 |
| M. tuberculosis (n=30) | miR.20 | 1.0000000 | 2.751453 | 0.5519 | 1.448 |
| M. avium (n=5) | miR.20 | 3.351521 | 0.827732 | 2.346 | 4.357 |
| M. kansasi (n=5) | miR.20 | 0.918965 | 0.102102 | 0.66 | 1.178 |
| M. simiae (n=5) | miR.20 | 2.271031 | 0.465163 | 1.702 | 2.84 |
| M. chelonae (n=5) | miR.20 | 2.823208 | 0.586672 | 2.108 | 3.539 |
| M. abscessus (n=5) | miR.20 | 2.955683 | 0.575207 | 2.252 | 3.66 |
| M. tuberculosis (n=30) | miR.29 | 1.000000 | 2.535365 | 0.5628 | 1.437 |
| M. avium (n=5) | miR.29 | 3.523421 | 0.579871 | 2.811 | 4.236 |
| M. kansasi (n=5) | miR.29 | 3.659534 | 0.661059 | 2.849 | 4.47 |
| M. simiae (n=5) | miR.29 | 1.180388 | 0.218416 | 0.9132 | 1.448 |
| M. chelonae (n=5) | miR.29 | 4.84966 | 1.213711 | 3.379 | 6.321 |
| M. abscessus (n=5) | miR.29 | 1.247874 | 0.257876 | 0.9328 | 1.563 |
| M. tuberculosis (n=30) | miR.155 | 1.0000000 | 1.104663 | 0.6854 | 1.315 |
| M. avium (n=5) | miR.155 | 1.373781 | 0.278359 | 1.033 | 1.714 |
| M. kansasi (n=5) | miR.155 | 0.65185 | 0.158526 | 0.4602 | 0.8435 |
| M. simiae (n=5) | miR.155 | 1.355692 | 0.282161 | 1.011 | 1.7 |
| M. chelonae (n=5) | miR.155 | 1.59555 | 0.265773 | 1.269 | 1.922 |
| M. abscessus (n=5) | miR.155 | 1.219381 | 0.160049 | 1.022 | 1.417 |
Table 2: The fold changes and Standard Division (SD) Mtb in comparison to NTM with the lower upper 95% Confidence Intervals (%CI) of mean.
Figure 3: The relative expression of exosomal miR-20a-5p, miR-29a, miR-let7e and miR-155 in Mycobacterium tuberculosis patients compared to nontuberculous
mycobacteria. Real-time PCR of exosomal apoptosis miR-20a-5p, miR-29a, miR-let7e showed significant statistics in non-tuberculous
mycobacteria compared to Mtb (p ≤ 0.0001). However, no significant analysis was determined for miR-155 in Ntb compared to Mtb. Data represent
mean and SD from 25 Ntb and 30 Mtb patients. Note:  Non-tuberculous mycobacteria
Non-tuberculous mycobacteria
Diagnostic power of serum exosomal apoptosis of miRNAs from Mycobacterium tuberculosis compared to healthy controls
Biomarker analysis of putative miRNAs; Hsa-miR-29a, Hsa-miR- let7e, Hsa-miR-155 and Hsa-miR-20a-5p was only assessed for Mycobacterium tuberculosis in comparison to control healthy subjects by performing ROC curve analyses. Statistical analysis was un- computed for NTM due to inadequate sample size. Up regulation of each miRNA might differentiate Mycobacterium tuberculosis from controls with AUC=0.73 (95% CI: 0.6041 to 0.8603, p<0.002) for Hsa-miR-let7e , AUC=0.69 (95% CI: 0.55 to 0.8, p<0.01) for Hsa-miR-20a-5p, for Hsa-miR-29a AUC=0.6 (95% CI: 0.45 to 0.74, p<0.1) , and significant differences was found for down regulation expression Hsa-miR-155 compared with healthy controls AUC=0.7 (95% CI: 0.61 to 0.87, p<0.001). Combinations of all miRNAs improved the AUC; a panel of Hsa-miR-20a-5p, Hsa-miR-29a, Hsa-miR-let7e and Hsa-miR-155 represents an AUC=0.86 (95% CI: 0.77-0.95, p<0.0001) vs. control subjects. (Table 3, Figures 4 and 5) .
| miRNA | AUC | P. value £ 0.05 | Youden’s index |
|---|---|---|---|
| miR-20a | 0.6933 | 0.01 | 0.37 |
| miR-29a | 0.6011 | 0.17 | 0.33 |
| miR-let7e | 0.7322 | 0.002 | 0.47 |
| miR-155 | 0.7456 | 0.001 | 0.47 |
| Combinations of miR-20a, miR-29a, miR-let7e, miR-155 | 0.86 | 0.0001 | 0.63 |
Table 3: Predictive values from AUC data of the ROC curve analysis for serum exosomal apoptosis of miR-20a-5p, miR-29a, miR-let7e, miR-155 separately and in combination with tuberculosis.
Figure 4: Receiver Operator Characteristic (ROC) curve analysis of serum exosomal miRNA (A) miR-20a-5p, (B) miR-29a, (C) miR-let7e and (D) miR- 155 level in patients with Mycobacterium tuberculosis. Receiver Operator Characteristic (ROC) curves were established using each microRNA expression value to determine the diagnostic power of the exosomal apoptosis miRNAs in MTB patients compared to healthy subjects. The Area under the Curve (AUC) with 95% CI and the P values were computed and presented for each ROC curve (miR-20a-5p: AUC=0.6933, p ≤ 0.01, miR-29a: AUC=0.6011, p ≤ 0.17, miR-let7e: AUC=0.7322, p ≤ 0.002, miR-155: AUC=0.7456, p ≤ 0.001). The optimal diagnostic point was determined at cutoff values with significant Youden’s index (sensitivity and specificity-1). (E) Combination of miR-20a-5p, miR-29a, miR-let7e and miR-155 with an AUC 0.86 and p ≤ 0.0001.
Figure 5: The expression level of serum exosomal miRNAs, (A) miR-20a-5p, (B) miR-29a, (C) miR-let7e, and (D) miR-155 of patients with Mycobacterium tuberculosis compared to healthy subjects reconstructed using mean -ΔΔCt. The expression of miR-20a-5p, miR-29a, miR-let7e overexpressed in the patients (-ΔΔCt: 1.38 p ≤ 0.03, -ΔΔCt: 0.95, p ≤ 0.30, -ΔΔCt: 1.23 p ≤ 0.01, respectively). Down regulation was found in miR-155 (-ΔΔCt: -1.038 p ≤ 0.006). (E) A combination of the miRNAs showed -ΔΔCt 2.48 with p ≤ 0.0001.
Mycobacteria could modulate the host immune response in different ways. One of these mechanisms is regulating the expression of diverse genes and their gene products which target various signaling pathways via miRNAs. These miRNAs were introduced as a class of small non-coding RNAs that consider key roles in post-transcriptional regulation of gene expression and have altered expression levels in various diseases [27, 28]. The current study indicates the up regulation level of Hsa-miRNAs 20a-5p, Hsa-miR-29a, and Hsa-miR-let7e in the serum of mycobacteria patients, while Hsa-miR-155 was down regulated in these patients. The diagnostic efficacy of the four miRNAs was improved using combined miRNAs (AUC=0.86, P ≤ 0.0001) in comparison with tested the miRNAs individually (AUC=0.6-0.74, P ≤ 0.01-0.001). One important part of the study is to assess the biomarker power of these selected miRNAs among NTM and Mtb vs. healthy controls that the expression of Hsa-miR-let7e, Hsa-miR-20a-5p, and Hsa-miR- 29a in Mycobacterium avium vs. Mtb was up regulated (P ≤ 0.01, P ≤ 0.0001, and P ≤ 0.0001, respectively). Also, expression of both Hsa- miRs let7e and Hsa-miR-20a-5p were increased in Mycobacterium abscessus vs. Mtb (P ≤ 0.0001 and P ≤ 0.002, respectively). Hence, as a pilot study, these results may introduce miRs let7e, Hsa-miR- 20a-5p, and Hsa-miR-29a as a potential diagnostic biomarker of Mycobacterium avium from Mtb infection. This is the same for Mycobacterium abscessus from Mtb infection by up regulation of Hsa- miRs let7e and 20a-5p, or overexpression of Hsa-miRs let7e and 20a-5p in Mycobacterium abscessus vs. Mycobacterium kansasi.
The present study conflicted with Guoliang Zhang's study on down-regulation of Hsa-miR-20a-5p that results in elevated cell apoptosis [23]. Guoliang Zhang reported that down-regulated Hsa-miR-20a-5p triggers cell apoptosis resulting in mycobacterial clearance by targeting JNK2 in human macrophages. They found down-regulation of 14 miRNAs in CD14+ monocytes of active pulmonary tuberculosis patients and miRNA-20a-5p was down- regulated after treatment by standard anti-tuberculosis regimen. The expression level of Hsa-miR-20a-5p was decreased in THP-1 monocyte cells after mycobacterial infection in-vitro. The study found that up-regulation of Hsa-miR-20a-5p increases mycobacterial survival and a decrease in the expression level of Hsa-miR-20a-5p facilitates macrophage apoptosis [23]. Up regulation of Hsa-miRlet- 7e and Hsa-miR-29a was found in Mycobacterium avium within human monocyte-derived macrophages resulted in to increase expression of caspase-3 and caspase-7 [29]. But, one contrasting study reports down-regulation of Hsa-miR-29 and induction of IFN-γ in NK cells and T cells of Mycobacterium bovis Bacillus Calmette-Guerin (BCG) [24]. This result indicates Hsa-miR-29 inhibition may have facilitated IFN-γ production by T cells and the expression of Hsa-miR-29 is influenced by Mycobacterium species- specific virulence. Concerning Hsa-miR let-7e, in 2019, Lei Feng and colleagues showed down-regulation of Hsa-miR let-7e in Knee OsteoArthritis (KOA) resulted to promote apoptosis and decrease autophagy. The study assessed Hsa-miR let-7e as a serum biomarker for KOA detection. The level of serum Hsa-miR let-7e was highly decreased versus controls. The expression level of proteins within the apoptotic pathway was enhanced in the cartilage of KOA rats. Circulating Hsa-miR let-7e may consider as a potential serum biomarker for KOA diagnosis [30]. Also, Krause and et al. in 2015 analyze Hsa-miR let-7e and 126 miRNAs in plasma for finding a biomarker of metabolic dysfunction in children aged from 10 to 12 years old. The subjects included 3325 school-age children. The plasma level of Hsa-miR let-7e was enhanced (˜3.4 fold) in subjects compared to the control group. An increase in plasma level of Hsa-miR let-7e shows a promising biomarker candidate for early detection of metabolic dysfunction in children with metabolic syndrome traits [31].
Our study shows that Hsa-miR-155 was down regulated in Mtb and NTM vs. healthy controls. Also, no statistical significance was determined in the expression level of Hsa-miR-155 in NTM vs. Mtb. Results conflict with Rajaram et al. [32] from another study [33] showing that virulent mycobacterial infection in human macrophages down regulates Hsa-miR-155 expression, but infection in murine macrophages up regulate Hsa-miR-155 expression. In comparison with virulent mycobacterium, infection of macrophages with avirulent mycobacteria species enhances Hsa-miR-155 expression level and increases TNF production. Moreover, one study has shown that Hsa-miR-155 was highly up regulated in Mycobacterium tuberculosis-infected murine RAW264.7 cells and bone marrow-derived macrophages [34]. Another study reported serum Hsa-miR-155 expression level was down-regulated concerning FOXO3 and TB-suppressing activity of cells [35], thus Hsa-miR-155 inhibits apoptosis of monocytes in patients with active TB through a negative regulation expression of FOXO3 [36]. Dysregulation of Hsa-miR-155 can also be associated with different cancer types other than mycobacteria. In 2021, Liang found overexpressed Hsa-miR-155 is accompanied by a poor prognosis of Acute Lymphoblastic Leukemia (ALL) in children and increases proliferation through targeting ZNF238 resulted to inhibit apoptosis [37]. However, Cerda and et al. in 2021 identified that down-regulation of Hsa-miR-155 is associated with metabolic syndrome (MetS) and cardiometabolic risk in obese patients (48 MetS and 32 non-MetS) [38]. Those debated results require more comprehensive research to a better understanding of the similarities and differences in human and murine macrophages study in correlation with miRNAs regulation [29].
A limitation of the current study is the inadequate sample size of NTM species. Hence, the diagnostic power of the exosomal apoptosis miRNAs was statistically un-computed in NTM cases and it was only analyzed for TB patients. Moreover, further studies with standardised methods are needed to analyse the use of these serum exosomal apoptosis miRNAs before and after use of anti- mycobacterial treatments, to better address the potential diagnostic power of serum-derived exosomal miRNAs in pulmonary tuberculosis and non-tuberculosis. Another significant limitation is additional control samples from patients who had infections that involved other bacteria must be included in further studies in order to show that these biomarkers candidates are only specific to mycobacteria. These putative miRNAs are not specific for only mycobacterial infection and the expression level might be up- regulated or down-regulated in different disease types including other infectious diseases. Further studies with standardized methods are required to represent the use of these serum exosomal miRNAs. miRNAs expression profiling may provide beneficial clues for pathophysiological studies and improve diagnosis, prognosis, and therapeutic measures.
This work was supported by the National Research Institute of tuberculosis and Lung Diseases (NRITLD), Shahid Beheshti University of medical sciences, Tehran, Iran.
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
The study conception was designed and approved by all authors. Data analysis management and material preparation were done by Alireza Javadi and Elaheh Ainy. Resources were prepared by Alireza Javadi and Payam Tabarsi. The study was supervised by Bahram Kazemi and Masoud Shamaei.
All participants provided written informed consent to participate in the study. Also, informed consent to publish data of the current study was obtained from all participants. All the techniques and methods were performed according to relevant guidelines and regulations. Also, the methods of the current study was approved by an institutional ethical board from Masih Daneshvari Hospital, Tehran, Iran (SBMU 1.REC1394.137).
The authors declare that there are no competing interests.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Javadi A, Shamaei M, Tabarsi P, Ainy E, Kazemi B, (2023) miR-20a-5p, miR-29a, miR-let7e, and miR-155 Serum Exosomal microRNAs Analyses as Potential Biomarkers in Pulmonary tuberculosis and Non-tuberculosis. Mycobact Dis.13:311.
Received: 02-Jan-2023, Manuscript No. MDTL-23-21726; Editor assigned: 05-Jan-2023, Pre QC No. MDTL-23-21726 (PQ); Reviewed: 19-Jan-2023, QC No. MDTL-23-21726; Revised: 26-Jan-2023, Manuscript No. MDTL-23-21726 (R); Published: 02-Feb-2023 , DOI: 10.35248/2161-1068,.23.13.311
Copyright: © 2023 Javadi A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.stricted use, distribution, and reproduction in any medium, provided the original author and source are credited.