Fungal Genomics & Biology
Open Access
ISSN: 2165-8056
ISSN: 2165-8056
Research Article - (2019)Volume 9, Issue 1
Background: Thecaphora frezii Carranza and Lindquist causes smut disease in peanut (Arachis hypogaea L.) resulting in up to 35% yield losses. Fungicides have shown ineffective in controlling the disease; whereas research on the molecular basis of that fungicide resistance has been hindered because of the lack of genetic information about T. frezii. The goal of this work was to provide molecular information about fungicide-target loci in T. frezii, including its mitochondrial genome (mitogenome) and critical nuclear-encoded genes.
Results: Here we report the complete annotated mitogenome of T. frezii, a 123,773 bp molecule containing the standard 14 genes that form part of mitochondrial complexes I, III, IV and V, 22 transfer RNAs, small and large subunits of ribosomal RNA, DNA polymerase, ribonuclease P, GII-reverse transcriptase/maturase, nine hypothetical open-reading frames and homing endonucleases (LAGLIDADG, GIY-YIG, HEG). In addition, we report the full-length cDNA sequence of T. frezii cytochrome b (cob) and cytochrome oxidase 1 (cox1) genes; as well as partial sequences of T. frezii succinate dehydrogenase (sdhb), ergosterol biosynthesis (Erg4), cytochrome P450 (cyp51), and beta tubulin (β-tubulin) genes, which are respective targets of strobilurins, quinone oxidation inhibitors, triazoles and beta-tubulin inhibitor fungicides commonly used in the peanut crop. Translation of cob and sdhb genes in this particular T. frezii isolate suggests potential resistance to strobilurin and carboxamide fungicides.
Conclusion: The mitogenome and nuclear-encoded gene sequences presented here provide the molecular tools to research T. frezii fungicide-target loci.
Mitochondrial genome; Pathogens; Groundnut; Carbon del mani; Fungicides
Currently endemic to Argentina, peanut smut is an emerging disease that constitutes a threat to the peanut industry around the world [1]. The etiological agent is the Basidiomycete fungus T. frezii, originally described on wild peanuts in Brazil in 1962 [1] and detected in the peanut production area of Argentina in the early 1990s [2]. Argentina is the seventh peanut producer in the world but is the first exporter in the peanut market [3]. T. frezii invades peanuts when the pegs touch the ground [4], and upon infection, seeds are replaced by a dark-brown powder of fungal teliospores (Figure 1) [5,6]. Without proper crop rotations T. frezii teliospores accumulate in soil, resulting in up to 51% of disease incidence [7] and up to 35% losses [8]. Much concern has spread throughout the peanut community, thus, in May 2017 the Australian peanut growers asked for a ban of peanut imports from Argentina [9], and in October 2017, the United States Department of Agriculture (USDA) - Animal and Plant Health Inspection Service (APHIS) restricted imports of raw peanut from Argentina and Brazil due to peanut smut [10]. Though peanut smut disease has not been reported in the United States, trade, travel and climate change may increase the risk of the disease being introduced and spread. Currently, United States is the fourth largest producer of peanut in the world (2.6 Mt) and Argentina is the eighth [11].

Figure 1: Symptoms caused by peanut smut, Thecaphora frezii, the dark powder is a mass of fungal teliospores developed inside the peanut pods after infection. Left: peanut pods still attached to the plant, Right: close view of infection.
Mitochondria in fungi participate in virulence, pathogenicity, survival and drug resistance, but they are essentially involved in energy metabolism [12]. Consequently, fungicides commonly target the energy-production protein complexes located in the inner membrane of the mitochondria (Figure 2) [13]. For example, diflumetorim (Pyricut) from the group of NADH inhibitors is a fungicide that targets complex I by inhibiting the NADH oxidoreductase activity [14]. Carboxamides (succinate dehydrogenase inhibitors, SDHI group) target complex II [15]. Strobilurins (Quinone oxidation inhibitors, QoI) target complex III, by inhibiting the electron transfer in the cytochrome bc1 and paralyzing ATP production [16]. Lastly, complex V is targeted by fentin hydroxide, or triphenyltin hydroxide [17], listed by the Fungicide Resistance Action Committee (FRAC) as an inhibitor of the oxidative phosphorylation (ATP synthase). Carboxamides and strobilurins are the most common fungicides used in the peanut crop along with triazoles and phtalonitriles, [1,18-20] (Supplementary Table 1).
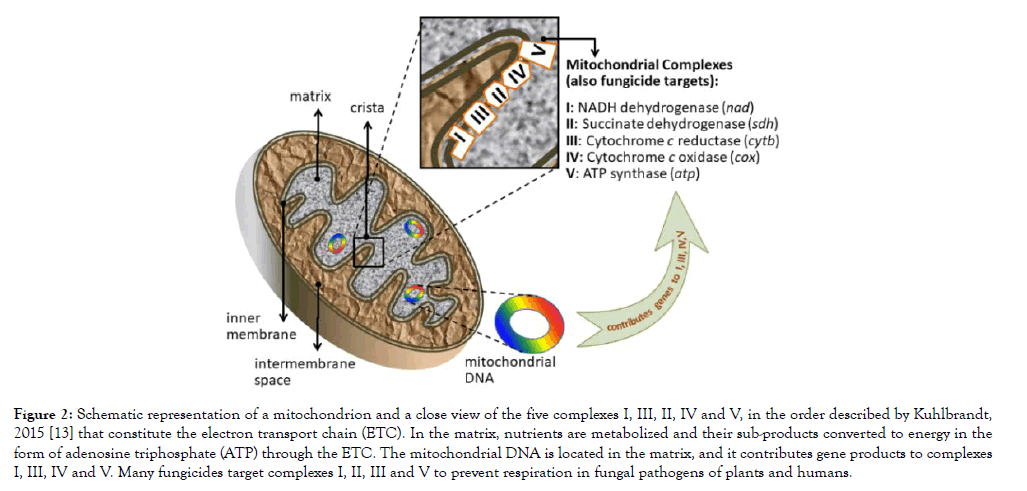
Figure 2: Schematic representation of a mitochondrion and a close view of the five complexes I, III, II, IV and V, in the order described by Kuhlbrandt, 2015 [13] that constitute the electron transport chain (ETC). In the matrix, nutrients are metabolized and their sub-products converted to energy in the form of adenosine triphosphate (ATP) through the ETC. The mitochondrial DNA is located in the matrix, and it contributes gene products to complexes I, III, IV and V. Many fungicides target complexes I, II, III and V to prevent respiration in fungal pathogens of plants and humans.
| Primers | Gene Sequence |
|---|---|
| TF_CYTB_Fw1 | GGAAATAATATGCGACTACTTAAATCACATCC |
| TF_CYTB_Rv1 | TACAGCTGTTCGTAGTAGAGGTGCTGTAGG |
| TF_COX1_Fw1 | ATGACTCGATGGCTTTTTTCAACAAACGC |
| TF_COX1_Rv1 | TAAGATTGACGTTAAAGCTACCCTCACTGGTG |
| TF_SDH_Fw | ATGACGATGTCGCTCTTCGCCGGCAACG |
| TF_SDH_Rv | ACGATCGCCAGCACCCTCACAGCTAGC |
| TF_Beta_Fw | CGCCGGTAACTACGAGGGCAGC |
| TF_Beta_Rv | TGGCGTATAGCTTCTTCAGG |
| TF_CYP51_Fw | CACCACCCTCACCCTACACGC |
| TF_CYP51_Rv | AGGACCACCATGGAAATCCGGC |
| TF_Erg4_Fw | CACTTTGAGTTCTGACTGACCGC |
| TF_Erg4_Rv | CTGACGATGCACCAGCAGGATCGC |
| T3 | ATTAACCCTCACTAAAGGGA |
| T7 | TAATACGACTCACTATAGGG |
Table 1: Primers used to amplify Thecaphora frezii genes that are target of fungicides, plus primers T3 and T7 that were added to their 5’ ends (forward and reverse, respectively).
Fungicide resistance frequently results from mutations in mitochondrial genes [21-23] and other fungicide-target loci [24-27]. To better plan strategies to control peanut smut it is essential to know the genetic makeup of T. frezii at those loci, but information about this species had been very limited. Until May 2019, there were merely 10 entries (607 to 722 bp long) for T. frezii in GenBank (NCBI database) corresponding to ribosomal RNA sequences and internal transcribed spacer (ITS) of T. frezii [28,29]. The objective of this work was to generate basic genetic information and molecular tools to investigate T. frezii and its response to fungicides. We report the complete annotated mitochondrial genome of T. frezii, as well as cDNA sequences of nuclear-encoded genes that are target of fungicides used in the peanut crop.
DNA extraction and sequencing
Peanut pods containing kernels with teliospores were collected from General Deheza (32° 45′ 08.54″S, 63° 46′ 06.5″W), Cordoba, Argentina, in 2016. Pods were surface disinfected in 0.26% NaOCl with continued agitation for 15 min, then 200 mg of teliospores were surface disinfected in 0.63% NaOCl for 5 min, rinsed twice with sterile distilled water and resuspended in 2 mL sterile distilled water [30]. Disinfected teliospores were plated on potato-dextrose agar (PDA) (Sigma-Aldrich, St. Louis, MO) supplemented with the extract of 50 g-L-1 peanut-seeds and incubated in the dark at 25°C [31]. DNA was extracted from the mycelial growth using the CTAB method [32], and identified as T. frezii by 18S ribosomal RNA sequencing [28]. DNA of an isolate identified as “A” was sent to the USDA-ARS-National Peanut Research Laboratory (NPRL) (Dawson, GA, U.S.A.), and the isolate was stored in the culture collection of the Instituto de Patología Vegetal (IPAVE) at the Instituto Nacional de Tecnología Agropecuaria (INTA), Cordoba, Argentina. Genomic DNA of isolate A was sequenced using Illumina HiSeq 2500 at LC_Sciences (Houston, TX) and at the USDA-ARS-Genomics and Bioinformatics Research Unit (GBRU) (Stoneville, MS). Reads from those runs were trimmed and assembled de novo at the NPRL using CLC_Bio Genomics Workbench 12, Qiagen (Redwood City, CA). No contaminating sequences were detected. A contig identified by BLAST analysis [33] as mitochondrial DNA was preliminary annotated with MAKER [34,35]. Additional open reading frames (ORFs) were predicted using Clone Manager Professional 9 (Sci-Ed Software, Denver, CO), and a map was built using Seqbuilder followed by GenVision in DNASTAR Lasergene 11 Core Suite (DNASTAR, Inc. Madison, WI). ORFs and hypothetical protein genes were translated by Clone Manager software and protein sequences subjected to search of motifs in Pfam and PROSITE databases using MOTIF (Kyoto University Bioinformatics Center, https://www.genome.jp/tools/motif/) to further characterize both predicted and intron encoded proteins. Mycelium preserved in RNAlater solution (Ambion, Life Technologies, Austin, TX) was used to extract RNA from the same T. frezii isolate using Direct-zol RNA MiniPrep Plus (Zymo Research, Irvine, CA), and two cDNAs were synthesized, one with oligo dT and the other with oligo DT+random hexamers using Superscript III reverse transcriptase (Invitrogen, Life Technologies, Carlsbad, CA).
Gene amplification and annotation.
To validate the annotation of T. frezii mitogenome, primers were designed to amplify transcripts of cytochrome b (cob) and cytochrome c oxidase 1 (cox1); both genes were PCR amplified from T. frezii cDNA libraries by Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Life Technologies, CA), using the following PCR conditions for amplification: 94°C 2 min; then 36 cycles of 94°C 30 s, 60°C 30 s, 68°C 1 min; then 68°C 5 min and kept at 4°C. The sequences of the universal forward primers of the T3 and T7 bacteriophages (Table 1) were added to the forward and reverse primers of the specific genes to increase the length of quality-sequencing obtained. PCR products were sequenced on SimpleSeq reactions at Eurofins Genomics LLC (Louisville, KY), the sequences were assembled using SeqMan Pro in DNASTAR Lasergene 11 Core Suite (DNASTAR, Inc. Madison, WI). Exon and intron annotation of cox1 and cob were revised according to the results, their corresponding proteins were translated using Clone Manager Professional 9 (Sci-Ed Software, Denver, CO) with the genetic code of yeast mitogenome [36].
Nuclear-encoded fungicide-target genes of T. frezii
We performed several iterations of mapping and de novo assembly and the final assemblies were screened by BLASTn in NCBI. Primers were designed to amplify nearly the full length of each gene from cDNAs of T. frezii. Gene coding regions were PCR amplified by the same conditions described for genes cob and cox1, using as template cDNA and DNA from T. frezii. Universal primers T3 and T7 were added to the 5’ end of forward and reverse primers, respectively, to increase the length of quality-sequencing obtained as indicated for cob and cox1 genes (Table 1). Putative T. frezii succinate dehydrogenase B subunit (sdhb), a lanosterol 14β-demethylase cytochrome P450 monooxygenase 51 (cyp51), ergosterol biosynthesis gene 4 (Erg4) and β-tubulin partial sequences were obtained by the described iterations of mapping reads, de novo assembly in CLC_Bio Workbench 11, and BLAST analysis. These sequences were submitted to the NCBI Database.
Thecaphora frezii mitogenome summary
T. frezii genomic DNA was sequenced. The total number of reads in two Illumina sequencing runs after trimming was 8,442,832 and 53,186,795, respectively, with an average length per read of 165 bp. After de novo assembly, one contig of 123,895 bp with a 10,664 x average coverage was identified as mitochondrial DNA by BLAST analysis. A 122 bp overlap of 5’ and 3’ ends allowed circularization into a 123,773 bp mitochondrial DNA molecule that constitutes the T. frezii mitogenome. The sequence was submitted to GenBank and has Accession number MH392473.
The T. frezii mitogenome contained genes identified as: seven units of nicotinamide-adenine dinucleotide reduced-form (NADH) reductase genes (nad) (1, 2, 3, 4, 4L, 5, 6); small ribosomal protein 3 (rpS3); partial large subunit of ribosomal RNA (rRNA); small subunit ribosomal RNA (rns); Ribonuclease P (EC 3.1.26.5) or ribozyme (rnpB); three units of ATP synthase subunit genes: atp6, atp8 and atp9; a putative DNA Polymerase (DNApol); three hypothetical proteins; another 7 open reading frames (ORFs); plus IEND, HEG, GIY genes, all represented in dark-blue arrows in Figure 3. Two ORFs (ORF255 and ORF186) were translated, and searches using MOTIF identified ORF 255 [70 amino acids (aa), pI: 10.13, MW: 7,592] as the N-terminal domain of a reverse transcriptase/maturase of group II (Expected value 5.4 × 10-16). ORF186 (186aa, pI:10.82, MW: 21,880) Group II intron maturase specific domain (E-value: 6 × 10-16), thus they were annotated as GII_MAT in the diagram. The mitogenome also had 23 transfer RNA (tRNA) that included all 20 amino acids plus two copies of transfer RNAs for methionine trnM (cat), arginine trnR (tcg, tct) and serine trnS (tga, gct), indicated as dark-red arrows in Figure 3. There were three subunits of cytochrome oxidase (cox), cox1, cox2, and cox3; only cox1 presented introns (light-green lines in Figure 3) for which MAKER predicted 14 introns and one more intron was identified by PCR amplification of cDNA and Sanger sequencing to a total of 15 introns. Also encoded in the T. frezii mitogenome was the cytochrome oxidase B gene (cob) (8,477 bp), which contained five introns, this was confirmed by PCR amplification and sequencing of cDNA. A total of 34,055 bp of the 123,773 bp T. frezii mitogenome (27.5%) corresponded to introns. In the reported orientation, T. frezii mitogenome had a 67.5% (A+T) and a 32.5% (G+C) content, color coded red/yellow for AT and blue/green for GC in Figure 3.
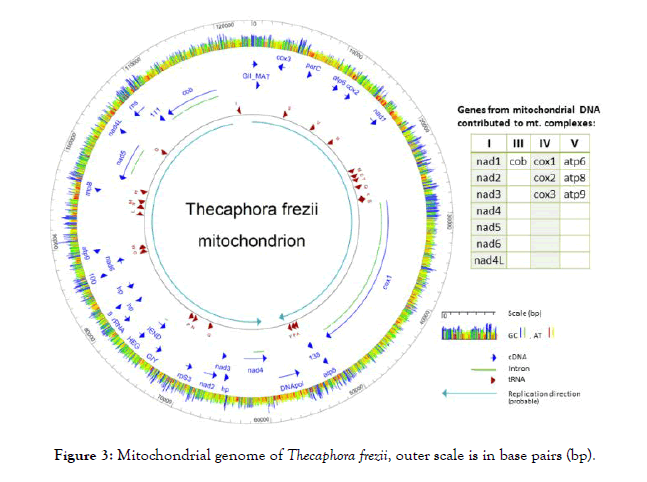
Figure 3: Mitochondrial genome of Thecaphora frezii, outer scale is in base pairs (bp).
The probable direction of replication of the molecule is indicated by two light-blue color arrows in Figure 3, and its origin was used as the start of the nucleotide sequence of the mitogenome. In the T. frezii mitogenome, we observed 19 type I introns, 3 type II introns, 6 proteins identified as endonucleases (LAGLIDADG, GIY-YIG, IEND, HEG). Other endonucleases were encoded within the introns of cox1 and cob genes [6 LAGLIDAG, 1 GIY-YIG, and 1 (RT)-RNA-dependent DNA Polymerase (RT_POL)], for clarity these were not included in the map annotation of Figure 3 but are shown separately in Figure 4. The reverse transcriptase (RT_POL) ORF found in cox1 was encoded in frame with the first exon of cox1 (overlapping the first 55 aa) and continuing within the first intron. These 895 aa proteins had a reverse transcriptase RVT_1 domain aa400-652 predicted in Pfam (Expected value (E) 1.9 × 10-33) and a Type II intron maturase at aa680-803 in Pfam (E 3.8 × 10-32). Also within cox1, Pfam predicted LAGLIDADG ORFs in introns 5 (E 1.5 × 10-18), 8 (E 2.9 × 10-8), 10 (E 1.4 × 10-4) and 14 (E 4.5 × 10-12), and a GIY-YIG (E 2.4 × 10-5) motif predicted by Pfam within intron 6 (Figure 4). Two open reading frames identified in Pfam as LAGLIDADG endonucleases were found in cob gene within introns 1 (E 7.3 × 10-14) and 3 (E 1.7 × 10-52), respectively (Figure 4).
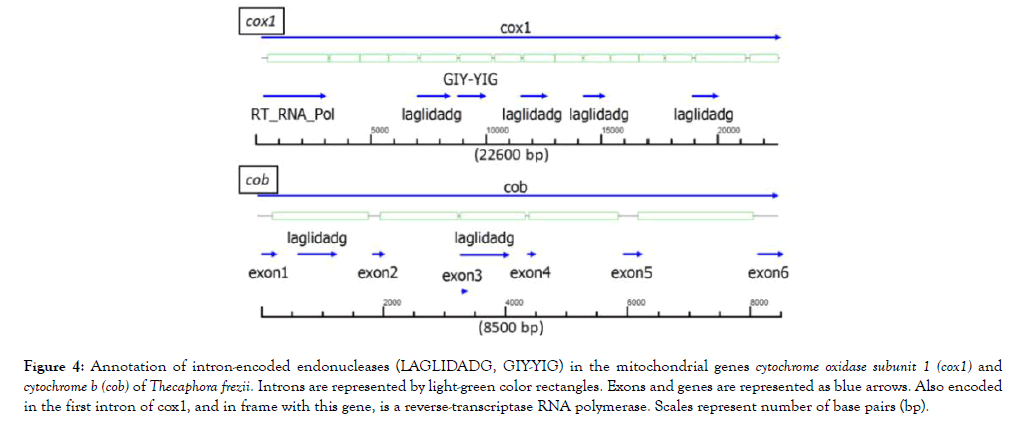
Figure 4: Annotation of intron-encoded endonucleases (LAGLIDADG, GIY-YIG) in the mitochondrial genes cytochrome oxidase subunit 1 (cox1) and cytochrome b (cob) of Thecaphora frezii. Introns are represented by light-green color rectangles. Exons and genes are represented as blue arrows. Also encoded in the first intron of cox1, and in frame with this gene, is a reverse-transcriptase RNA polymerase. Scales represent number of base pairs (bp).
cob and cox1 genes
Primers used to amplify cob and cox1 genes from cDNAs (oligo dT, oligo dT + random hexamers) are listed in Table 1. The sequences of the universal primers T3 and T7 were added as indicated before (Table 1). The presence of long introns in the cob gene of T. frezii (Figure 4), made its total length 8,478 bp, though its 1,200 bp transcript encoded only 400 aa. The cDNA sequence of the cob gene was translated using the yeast-mitochondrial genetic code [36] reading UGA codons 273, 349 and 351 as tryptophane (W) instead of stop codons, underlined in Figure 5. The translated COB protein had molecular weight MW: 45,989.2, isoelectric point (pI): 8.14. This sequence was submitted to GenBank (MH603949).
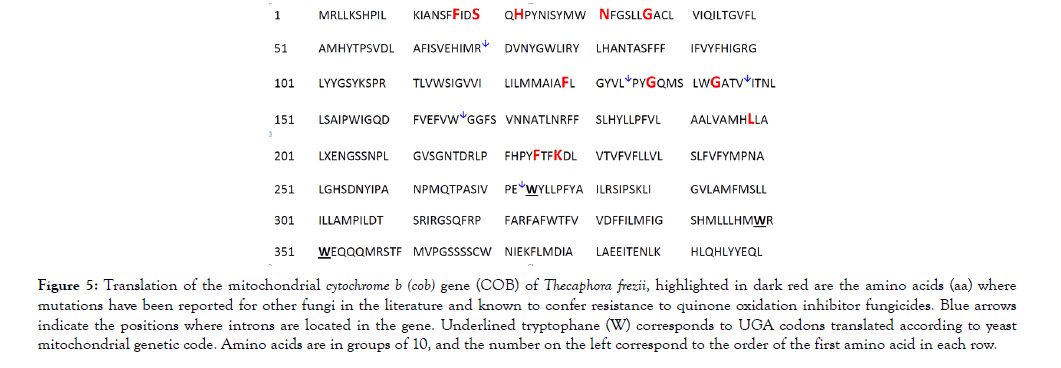
Figure 5: Translation of the mitochondrial cytochrome b (cob) gene (COB) of Thecaphora frezii, highlighted in dark red are the amino acids (aa) where mutations have been reported for other fungi in the literature and known to confer resistance to quinone oxidation inhibitor fungicides. Blue arrows indicate the positions where introns are located in the gene. Underlined tryptophane (W) corresponds to UGA codons translated according to yeast mitochondrial genetic code. Amino acids are in groups of 10, and the number on the left correspond to the order of the first amino acid in each row.
The total length of T. frezii cox1 gene was 22,731 bp, however, the cDNA was only 1,586 bp. Two variants of cytochrome oxidase 1 transcripts of cox1 gene were observed after PCR amplification of T. frezii cDNA, sequencing and translation. These were named TF_ COX1_v1 and TF_COX1_v2 in Figure 6, submitted to GenBank as MH603947 and MH603948. Both had 528 aa and pI: 6.39, though they had slightly different molecular weight (58,080.3 and 58,060.2, respectively). Five aa differences were observed for these two molecules between positions aa400 and aa495, predicting protein structural change according to the model of Garnier-Osguthorpe-Robson (Figure 6) [37]. The five amino acid changes were L400F, P451A, L466P, P490A and S495T. In the predicted model, TF_COX1_v2 has a β-sheet instead of turn at aa490-491; a β-sheet instead of a helix at aa462-466; an extended helix at aa449-451 not present in TF_COX1_v1; and an extended turn at aa399-400 in TF_COX1_v2 that is not present in TF_COX1_v1, (Figure 6).
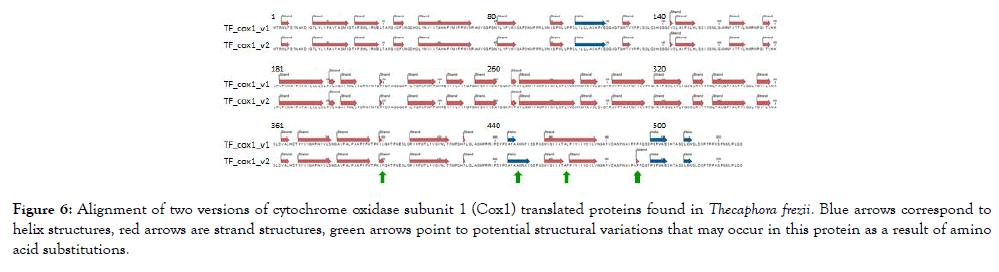
Figure 6: Alignment of two versions of cytochrome oxidase subunit 1 (Cox1) translated proteins found in Thecaphora frezii. Blue arrows correspond to helix structures, red arrows are strand structures, green arrows point to potential structural variations that may occur in this protein as a result of amino acid substitutions
Nuclear-encoded fungicide-target genes
Several iterations of mapping and de novo assemblies were performed searching T. frezii sequences for nuclear-encoded genes that are target of fungicides. The T. frezii sequences submitted to GenBank corresponded to a 1,648 bp putative succinate dehydrogenase subunit B (sdhb) (MH603943), 1379 bp of β-tubulin gene (MH603944), 1,595 bp of cytochrome P450 monooxygenase 51 (cyp51) gene (MH603946), and 1,865 bp of ergosterol biosynthesis gene 4 Erg4 (MH603945). SDHB of the sequenced T. frezii presented aa arginine (R) at the position 275, position that corresponds to 272 in SDHB of B. cinerea (Pers.) (Botryotinia fuckeliana) or 257 in SDHB U. maydis, Primers reported here can amplify a 1,379 bp of the β-tubulin gene in T. frezii including the region that corresponds to amino acids 198 and 200. In the T. frezii isolate sequenced mutations E198A and F200Y normally associated to resistance to benzimidazole fungicides were not observed,
Fungicide target genes
Mutations, usually single-nucleotide polymorphisms (SNPs), in fungal plant-pathogen genes encoding fungicide targets, have been often the cause of developed resistance to strobilurins (QoI), carboxamides (SDHI) and benzimidazole fungicides [21-23,27]. Therefore, monitoring such mutations in field-collected samples has been shown essential to determine fungicide-treatment programs in crops [27,38]. In T. frezii, the basic research to discover potential mutations in relation to fungicide resistance has not been possible, because of the lack of genetic information about this pathogen. The genome of Thecaphora thlaspeos, a related species that invades Brassicaceae plants, was recently published [39]. This will be a valuable resource to help accelerate research on T. frezii. Here we report the complete mitogenome of T. frezii and provide tested primer sequences to amplify mitochondrial and nuclear genes that are target of common fungicides used in the peanut crop. Examples of mutations related to fungicide resistance are mentioned for the sole purpose of illustrating their relevance; this is not a complete review of the mutations reported in the literature for fungicide resistance.
Strobilurins, carboxamides and triazoles are the most common fungicides used to control peanut diseases [1,18-20]. In peanut smut, however, various combinations of the mentioned fungicides have been less than 50% effective even when used at high doses [40,41]. It is known that mutations in genes encoding fungicide targets of pathogenic fungi are often the cause of developed resistance to strobilurins (QoI), carboxamides (SDHI) and triazole fungicides [21-23,27]. Nevertheless, research on the molecular basis of T. frezii fungicide resistance has not been possible due to the lack of genetic information. Here we report the complete mitogenome of T. frezii, (accession MH392473), and provide molecular tools to study fungicide target genes such as sdhb (MH603943), cytB (MH603949), Erg4 (MH603945), cyp51 (MH603946) and β-tubulin (MH603944). Some of the sequences in this isolate indicated potential resistance to strobilurins and carboxamides.
Cytochrome B: Here we report the complete sequence of DNA and mRNA of the cytochrome B of T. frezii, MH603949. Strobilurins fungicides target Cyt B in the mitochondrial complex III, and strobilurins account for approximately 25% of the global market of fungicides [42]. At least eight amino acid substitutions in Cyt B are known to confer resistance to strobilurins in other fungi [21,43]. Thus, the monitoring of mutations in the Cyt B of field samples of fungal pathogens has been essential to modify fungicide-treatment programs in other crops [27,38]. Since the positions of Cyt B mutations reported in other fungi correspond in T. frezii COB to different exons in a gene that is 8,478 bp, here we propose the use of cDNA and primers provided in Table 1 as a mean to capture all the potential mutations in this gene. The T. frezii isolate sequenced in this study presented the aa phenyalanine (F) at position 17 of the quinone reduction site (Qi) in Cyt B. Substitution I17F has been linked to Diuron and Ilicicolin H resistance in other fungi [43]. Other aa substitutions in Cyt B that normally result in fungicide resistance are F129L, G137R and G143A [21,43] but these were not observed in the T. frezii isolate sequenced.
Succinate dehydrogenase b: In most eukaryotes including fungi, all the genes that form the mitochondrial complex II, one being succinate dehydrogenase, are encoded in the nucleus of the cell [44]. The isolate of T. frezii sequenced here, presented aa arginine (R) at position 275 of SDHB protein. Presence of aa arginine at the corresponding position of SDHB in Botrytis cinerea is known to confer resistance to SDH inhibitors Boscalid and Fluopyram [45]. Thus, it is highly possible that the T. frezii isolate sequenced may have resistance to carboxamides.
Cyp51, Erg4 and Nad. Triazole fungicides target the ergosterol biosynthesis pathway by inactivating either the lanosterol 14 α-demethylase cyp51 gene (member of the P450 family, Erg11 in yeast) [46], or the ergosterol biosynthesis gene 4 (Erg4) [47], both nuclear-encoded genes. Mutations in these two genes, cyp51 gene [24,25] and Erg4 [47], or in the NADH oxidoreductase subunit of mitochondrial complex I are known to confer resistance to azole fungicides [24]. Here we report partial cDNA sequences of T. frezii cyp51, Erg4, and the nad mitochondrial genes of T. frezii that form the core of complex I. This information could be used as a starting point to research triazole resistance in T. frezii. However, additional mechanisms have recently been described as involved in azole resistance, such as a calcium transporter, transcription activation and a saccharopine dehydrogenase [48].
Beta tubulin: Primers reported here can amplify most of the β-tubulin gene of T. frezii, a 1,379 bp, including the coding region corresponding to amino acids 198 and 200. Mutations E198A and F200Y in the β-tubulin gene are known to confer resistance to benzimidazole fungicides in Monilinia fructicola and Botrytis cinerea [26,27]. The β-tubulin of T. frezii had high homology (89%) to Anthracocystis flocculosa PF-1 (XM_007882922.1) followed by 87% identity to the β-tubulin gene of Kalmanozyma brasiliensis (XM_016438208.1). No evidence of potential fungicide resistance was observed at positions 198 or 200 in the T. frezii isolate sequenced.
Mitogenome: endonucleases, gene orientation and heteroplasmy
In the mitochondrial genome of T. frezii, genes were oriented in two opposite directions away from the location of GII_MAT (Figure 3). In general, mitochondrial genes can all have the same orientation as for example in Rhizoctonia solani (NC_021436) [49], or they can be in opposite orientation as in Phlebia radiata Fr [50]. GII_MAT is a Group II transcriptase/maturase (GIIM) required in vivo for the splicing of group II introns, GIIM are mobile elements with multiple catalytic activities [51]. We speculate that the location of GII_MAT can be the origin of mitochondrial DNA replication.
BLAST analyses of T. frezii sequences showed homology to Anthracocystis flocculosa [Traquair et al.] Piatek et al., a fungus in the order Ustilaginales; though T. frezii is in the order Urocystidales. This probably occurred because currently there are 595 entries for Ustilaginales for each entry of Urocystidales. The T. frezii mitogenome sequenced had 52 genes in addition to intronencoded homing endonucleases. A 27.5% (34,055 bp) of T. frezii mitogenome corresponded to introns, that is the equivalent to twice the size of the entire human mitogenome (16,569 bp) which encodes 37 genes [52]. Fungal mitogenomes vary significantly in size, for example 23,743 bp in the ascomycete Zasmidium cellare [53] and 235,849 bp in Rhizoctonia solani [49]. Despite having similar number of genes, mitogenome sizes are usually expanded by homing endonucleases. For example, in the 203 Kb mitogenome of Sclerotinia borealis, more than half (125 Kb) correspond to introns, most of them containing homing endonucleases (LAGLIDADG and GIY_YIG) [54]. LAGLIDADG endonucleases are encoded within introns type I, type II, archeal introns and inteins to promote lateral transfer [55]. The T. frezii mitogenome contained 10 homing endonucleases (LAGLIDADG, GIY-YIG, HEG, IEND), 7 of them encoded in introns of cob and cox1 genes. Whereas the role of these endonucleases is not completely understood; fungal transcriptome analysis of the effect of a new fungicide, JS399-19 on Fusarium graminearum showed downregulation of only few proteins, among them was a LAGLIDADG [56]. Thus, it would be interesting to further characterize the role of these endonucleases in T. frezii in relation to fungicide response.
In the sequenced isolate of T. frezii the total length of cytochrome oxidase 1 gene (cox1) was 22,731 bp due to the presence of 15 introns encoding endonucleases. This gene in T. frezii showed heteroplasmy, that is two variants with possible structural differences, within their C-terminus as predicted by Garnier’s model [37]. The C-terminal domain of Cox1 regulates the Cox1 synthesis by modulating interactions between proteins of the mitochondrial complex IV [57]. Until recently, mitochondrial heteroplasmy in Basidiomycota was considered transient or non-existent, but in 2017, heteroplasmy of Cox1 was reported for the edible mushroom Thelephora ganbajun [58-60].
We have sequenced and annotated the mitogenome of T. frezii, as well as several fungicide target genes outside the mitogenome, these can be used to study the potential fungicide resistance in this pathogen. Amino acids predicted in COB and SDHB proteins indicated potential resistance to strobilurin and carboxamide fungicides. T. frezii cox1 and cob genes could be also useful for diagnostic application, as these genes have been successfully used for genetic-barcoding identification in other species.
Cazon, Conforto, Paredes, Rago: isolation and molecular identification of the fungal pathogen; Lamb, Soave, Buteler: leaders and coordinators in the international agreement for research on peanut smut; Scheffler, Duke, Simpson DNA sequencing; Arias: data analysis and wrote the manuscript, Massa and Sobolev run MAKER for annotation and made editorial comments.
This work was feasible thanks to the support of USDA-ARS project number 6604-21000-004-00D and 6044-21000-004-00D, National Peanut Research Laboratory, Dawson, Georgia, U.S.A., the collaboration of the Instituto Nacional de Tecnologia Agropecuaria (INTA), Cordoba, Argentina, and Fundacion Mani Argentino, Cordoba, Argentina. The authors would like to thank Ms. Valerie Orner at USDA-ARS-NPRL for her technical assistance.
Citation: Arias RS, Cazon LI, Massa AN, Scheffler BE, Sobolev VS, Lamb MC, et al. (2019) Mitogenome and Nuclear-encoded Fungicide-target Genes of Thecaphora frezii - Causal Agent of Peanut Smut. Fungal Genom Biol. 9: 160. doi: 10.35248/2165-8056.19.9.160
Received: 18-Jun-2019 Accepted: 21-Aug-2019 Published: 28-Aug-2019
Copyright: © 2019 Arias RS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.