Journal of Food: Microbiology, Safety & Hygiene
Open Access
ISSN: 2476-2059
ISSN: 2476-2059
Research Article - (2018)Volume 3, Issue 2
This study aimed to explore the dominant fecal microbiota and fecal short chain fatty acid metabolites in Egyptian adults. The impact of a three week dietary intervention with fermented sour sobya (SS) providing daily 1.56 ± 0.22 and 1.79 ± 0.14 billion colonies forming units of Lactobacilli and yeasts, respectively on the fecal microbiota and metabolites in healthy adults were also investigated. The abundances of 12 bacterial taxa were characterized before and after three-week daily intake of SS by quantitative real time PCR. Fecal short chain fatty acids were analyzed in parallel with gas chromatography. The intervention with SS led to more than 46-fold average increase in genome numbers of Lactobacillus rhamnosus, potential probiotic present in SS. Fecal Enterobacteriaceae and its genus Escherichia, which are often associated with pathogenic traits and inflammation, were reduced significantly following the three week intake of SS. The intake of SS was also associated with significant increase in fecal short chain fatty acids including butyrate. These data provide evidence of unique beneficial effects for the use of microbiome-based therapies such as fermented sour sobya in clinical and molecular nutrition and medicine.
Sour sobya, Egyptian adults, Gut microbiota, Short chain fatty acids, Lactobacillus rhamnosus , Enteric bacteria
CFU: Colony Forming Units; LAB: Lactic Acid Bacteria; MRS: Man Rogosa Sharpe Medium; SCFA: Short Chain Fatty Acids; YPD: Yeast Extract Peptone Dextrose Medium
The human gut microbiome contains a large number of bacterial cells and the bacterial cells harboured within the human gastrointestinal tract (GIT) were estimated to outnumber the host’s cells by a factor close to 1:1 [1] and in other studies 10:1 [2]. The gut microbiota is involved in basic human biological processes, including modulating the metabolic phenotype, regulating epithelial development, and influencing innate immunity [3-6]. The typical gut microbiome of an adult is dominated by the phyla Bacteroidetes and Firmicutes, with lesser numbers representing Proteobacteria, Actinobacteria, and Archaea [7]. Results from 16S rRNA gene-based microbiota profiling demonstrate that different long-term diets are associated with distinct gut microbiomes [8-10]. For example, both quantitative and qualitative changes of mucosal and fecal gut microbiota were reported in response to high intakes of dietary fats and refined sugars that are typical of a Western diet [11,12]. These changes can lead to disruptions of the community ecology of the microbiome with implication in diseases, particularly those involving systemic or localized inflammation [12-14].
With the recognition of adverse effects of diets rich in processed foods on the human gut microbiome [15,16] incorporation of probiotic bacteria and functional fermented foods into human consumption elicits an increasing interest [17-20]. Probiotic microorganisms provide benefits to the human host by modifying the composition of gut microbiota; enhancing resistance to potential pathogens, via competitive adherence to the mucosa and epithelium; competition for nutrients and the production of anti-microbial substances; strengthening gut epithelial barrier function; preserving epithelial barrier, decreasing abundance of pathogenic bacteria, and promoting a healthy intestinal immune system [21-24] and can reduce the risk of upper respiratory tract infections [25,26]. Probiotic dosages are measured in Colony Forming Units (CFUs), which express the live bacterial organisms provided in each dosage and probiotic foods must contain at least one billion (1 × 109) live colony-forming units of a recognized probiotic species per serving (e.g., active probiotic cultures). A wide range of dosages for Lactobacillus sp. and other probiotics have been studied in clinical trials and most studies examined dosages in the range of 1 to 20 billion cfu per day. More than 10 billion cfus per day in adults were associated with a more significant study outcome [27,28]. Synergistic multi-strain and multi-species probiotics were reported to be more effective over mono-strain probiotics [29-31] more effective at inhibiting pathogens than singlestrain probiotics, as the pathogen is thereby exposed to multiple forms of antagonism .Wide variation exists in the dosages provided by different probiotics than with almost any other dietary supplement used today [32-34]. The wide variation in the dosage has to do with the different forms of probiotics available today, individual product viability and intended product usage [35]. Probiotics are generally sold as capsules or are incorporated into food. Therapeutic fermented dairy products such as Danactive, which contains 10 billion CFUs of L. casei DN-114 001 per serving, and Activia, which contains about 5 to 10 billion CFUs of B. animalis DN-173 010 per 4-oz (113.4 g) container, are currently available. Consistent identification and concentration of the probiotic microorganism(s) must be clearly indicated on the label in order to avoid misunderstanding and misuse of the probiotic product. Because some labels are unreliable, precautions should be undertaken to recommend specific brands known to be of reasonable quality [36]. Another important aspect that might to be observed is related to the viability of the microorganism(s) at the time of consumption; often the number of probiotic bacteria found in the products was below the one declared. Probiotic supplement manufacturers will overdose their products so they provide a minimum number of viable CFUs through the end of their shelf-life [37] or by introducing the technology of microencapsulation to improve the shelf-life of probiotic bacteria and to ensure a consistent dosage is provided throughout the duration of the probiotics viability [38]. Popular commercial probiotic cultures which are available in global markets include Lactobacillus rhamnosus HN001 (DR20) marketed by Danisco (Madison,WI , USA), L rhamnosus R0011 marketed by Institute Rosell (Montreal, QC, Canada), and Saccharomyces cerevisiae (boulardii) marketed by Biocodex.
The Food and Agricultural Organization of the United Nations encouraged more extensive studies on traditional fermented foods because the acid-producing organisms in these products may prevent “fouling” of the large intestine and thus lead to an increased life span of the consumer [39]. Food fermentations are ancient processes that date back to the introduction of agriculture and animal husbandry and are estimated to make up approximately one-third of the human diet [40] and play essential roles in enhancing the stability, quality, flavor, and texture of human food production practices that entailed the recycling of isolated microbial communities in the presence of abundant agricultural food sources [41]. Lactic acid bacteria (LAB), Bifidobacteria and bread yeasts are the dominating environmental microorganisms [42] that enter the GI tract. During fermentation, the enzymatic activity of the raw material, colonic extraction of nutrients and the metabolic activity of microorganisms modify the food constituents, synthesizing metabolites and proteins, and providing living microorganisms to the gastrointestinal (GI) tract [43].
Artisanal fermented cereal grains are made from different types of cereals as maize, sorghum, millet, rice or wheat under the catalytic action of lactic acid bacteria and yeasts [44-46]. The methodologies are based on serial inoculation in a process known as back-slopping andresults in specifically organoleptic and health-beneficial properties [47]. LAB comprise a significant component of the human gut flora and have several beneficial roles in the probiotic potential of lactic acid bacteria and this group typically constitutes around 1% of the fecal bacterial population [48]. Traditional food fermentations are elegantly simple in that they generally required very few ingredients and fermented cereals have been increasingly recognized as functional foods with beneficial properties, preparation and processing. Although some fermentations contain only few dominant taxa, strain differences and population dynamics during process can be remarkably complex [47].
In some foods, even minor alterations to species diversity or numbers can result in significantly different food products and variations in quality and organoleptic properties. Therefore, a microbial composition with temporal and spatial stability and resilience results in consistent fermentations and process conditions those are necessary to produce high quality food. Recent studies have explored the microbial diversity of numerous fermented food types.
It is increasingly understood that some fermented foods also promote human health in ways not directly attributable to the starting food materials. That is, the outcomes of fermentation, and the contributions of microbes, in particular, can provide additional properties beyond basic nutrition. Recent human clinical studies on fermented foods support this possibility and related the intake of fermented food and improvement in non-communicable chronic diseases, such as type -2 diabetes Mellitus [49,50].
Sour sobya (SS) is fermented cereal grains milk porridge prepared by inoculation of wheat [45] or rice [51] with non-conventional diverse lactic acid bacteria (LAB) and mixed yeasts species [45]. Although only a limited number of clinical studies on fermented foods have been performed, scientific based evidences demonstrated that intake of SS reduced risk of cardiovascular disease (CVD) [52], intestinal permeability [53], stimulated the anti-oxidant capacity among healthy adults [54] and had in vitro anti-microbial activities against seven virulent enteric pathogenic bacteria [55].
These benefits might extend to immediate physiological responses, a possibility recently indicated by the finding that fermented milk consumption reduced muscle soreness induced by acute resistance exercise [56] and prevented the onset of physical symptoms in students under academic examination stress [57].
Currently, there is no data available on the gut microbiome of Egyptian children. The objective of the present study was to quantify the abundances of 12 common taxa of the human gut microbiome among Egyptian adults and to report changes in fecal microbiota following intake of fermented sour sobya.
Study design and participants. A randomized controlled trial was conducted on 14 healthy Egyptian volunteers with mean age of 24.4 ± 0.7 years and BMI average of 23.7 ± 0.9 kg/m2. The study protocol received approval from the National Research Center Medical Research Ethics Committee, Egypt (Registry number 16-422). The protocol was fully explained to all participants and written informed consent was obtained before participation in the trial. Exclusion criteria were defined as any symptoms that were likely related to a digestive tract disease and/or intake of antibiotics during three months prior to the start of the trial. Participants were asked to maintain their normal diet, but to abstain from consumption of probiotic-containing products or fermented dairy products. After two weeks washout period, the participants were divided randomly into two equal-sized groups. The first group (designated “SS group”) consumed daily 170 g SS, while the second group (designated “control group”) served as a control and did not receive any supplement.
Sour sobya is a commercial fermented sweet porridge (Elrahmany, SayedaZeinab, Cairo, Egypt) and was purchased in 500 g packages twice weekly from the same store and stored in the refrigerator. The supplement was delivered daily in 170 g portions providing 4.4 ± 0.62 and 1.79 ± 0.14 billion cfu of lactic acid bacteria and yeast, respectively. The dietary intervention study continued for 21 days and the volunteers were asked to collect voided stool samples in sterile containers at day 0 and day 21, which were saved at -70°C for subsequent analysis.
Dietary assessment
The participants completed food diaries three times per week (including two weekdays and one weekend day). Dietary intake was calculated using a computer aided nutritional analysis program (Nutri Survey, Seoul, Korea). Daily macronutrient consumption was calculated using a diet software program based on the type and quantity of each ingredient consumed, and Egyptian food composition tables.
Microbiological testing
The enumeration of the viable bacteria in the SS suspensions was carried out by plating serial dilutions of SS from 1/1000 to 1/107on the selective deMan, Rogosa and Sharpe Agar (MRS, Biolab, UK) lactobacilli on MRS agar (Difco, Sparks, MD, USA) supplemented with 3% ethanol and 0.5% cycloheximide to inhibit yeast growth [59]; in an anaerobic environment using anaerobic gas packs (Oxoid, Unipath Ltd, Basingstoke, UK), capable of producing 1800 cm3 hydrogen and 350 cm3 carbon dioxide. The plates were incubated at 30°C and examined after 5 days for the bacterial growth and the counts were expressed as cfu g-1 of SS. Colonies (Gram positive isolates) from the highly diluted plate were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) protein analysis [60]. Preparation of the bacteria isolate was carried out according to the ethanol/formic acid extraction protocol [61]. The MALDIBioTyper (BrukerDaltonics, Leipzig, Germany) was used for the analysis. The results were expressed by MALDI-BioTyper matching scores (ranging from 0.000 to 3.000), which indicated the similarity of the unknown MALDI-TOF MS profile to available profiles in the MALDI-BioTyper 3.0 software database.
The yeasts in SS were enumerated on acidified YEPG agar (yeast extract 10 g; peptone 20 g; glucose 20 g; agar 15 g per liter) which had been adjusted to pH 3.5 by the addition of sterilised 1N after being autoclaved [62]. The plates were incubated at 30°C for 72 h and colonies were enumerated and the yeast isolates were identified with API 50 CHL test (BioMe`rieux, Marcy-l’Etoile, France) following instructions of the manufacturer.
Fecal DNA extraction
Fecal specimen (150 mg) was promptly removed from the frozen sample and extraction was completed by the ZR fecal DNA isolation kit (DNA MiniPrep™ kit (Catalog No. D6010, Zymo Research, Ohio, USA) under simultaneous multi-directional beating in a FastPrep 24 instrument (MP Biomedicals, Ohio, USA) for 40 s . The extractions and purification were done following the manufacturer‘sprotocol based on published technique [63].
Genomic DNA quantification was based on the absorbance measurement at 260 nm wavelength using Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Genomic solution containing 50 μg ml−1 of double-stranded DNA has an OD 260 of 1.0 and was stored at -70°C. The DNA concentration was adjusted to 10 ng μl-1on the day of amplification.
The following twelve pure bacterial groups and species were obtained from the Deutsche Sammlung von Mikroorganismen (Braunschweig, Germany) or the American Tissue culture collection (ATCC)and were used for bacterial identification : Bacteroides thetaiotaomicron DSM 2079, Bifido bacterium ATCC 15707T, Bifidobacterium longum NCC 2705, Blautia product DSM 2950, Clostridium butyricum DSM 10702, Clostridium leptum DSM 753, Enterobacteriaceae DSM 30083T, Escherichia coli K-12 MG1655, Faecalibacterium prausnitzii DSM 17677, Lactobacillus DSM 20079T17, Lactobacillus rhamnosus ATCC 7469, Prevotella melaninogenica DSM 7089. The bacterial cultures were grown anaerobically in an atmosphere of 80%, N2-20% CO2 at 37°C. The purity of the cultures was checked by inspecting the colony morphology after anaerobic growth on BHI medium and cellular morphology with gram staining. The cells were suspended in buffer, centrifuged and the pellets (approximately 109 bacterial cells) were used for the extraction and purification of DNA using RTP Bacteria DNA Mini Kit (Invitek, Berlin; Germany). The extracted genomic DNA was stored at -70°C until used.Just before measurement, dilution series from 2.0 × 1010-2.0 × 102 copies/μl PCR reaction were done.
The sequences of each primer pair targeting 16S rRNAbacteria group or species are listed in Table 1 and the universal 16S gene PCR primer set (63F/1387R) was used to quantify the total bacteria. The housekeeping gene groEL HSP60 (EuroFins MWG Operon (Ebersberg, Germany) was used in the case of Bifidobacteria, due to its superior characteristics based on previous experiences [64].
| Target taxon | Primera | Primer sequences | Amplicon size | Annealing T (°C) | Reference |
|---|---|---|---|---|---|
| Bacteroides spp. | Bact-F | TGTGACTGCCGGTGCAAGCC | 192 | 71 | Slezak [90] |
| Bact-R | ACTTTGCGCATAGCGTCAGCA | ||||
| Bifidobacterium spp. | Bif-F | CTCCTGGAAACGGGTGG | 547 | 55 | Matsuki [91] |
| Bif-R | GGTGTTCTTCCCGATATCTACA | ||||
| Bifidobacterium longum | Blon-F | CGGCGTYGTGACCGTTGAAGAC | 257 | 70 | Junnick, Blaut [64] |
| Blon-R | TGYTTCGCCRTCGACGTCCTCA | ||||
| Blautiaproducta | Bla-F | AACCTGGCAGCAGGCGCTAAC | 149 | 71 | Slezak [90] |
| Bla-R | TCATCGCCTGCGGAGATAGCTG | ||||
| Clostridium butyricumb | Cbut-F | AGTAGCTGTTGAAAAGGCAGTTGAAGA | 99 | 71 | Slezak [90] |
| Cbut-R | TCAGCAGCAGAAATAGCAGCAACTA | ||||
| Clostridium leptumgroup | Clep-F | GCACAAGCAGTGGAGT | 250 | 50 | Matsuki [92] |
| Clep-R | CTTCCTCCGTTTTGTCAA | ||||
| Enterobacteriaceae | Ent-F | CATTGACGTTACCCGCAGAAGAAGC | 500 | 63 | Bartosch [93] |
| Ent-R | CTCTACGAGACTCAAGCTTGC | ||||
| Escherichia coli | Ecol-F | GGCTATCATCACTGAAGGTCTG | 100 | 67 | Dumonceaux [94] |
| Ecol-R | TTCTTCAACTGCAGCGGTAAC | ||||
| Faecalibacterium prausnitzii | Fpra-F | CCATGAATTGCCTTCAAAACTGTT | 142 | 71 | Sokol [95] |
| Fpra-R | GAGCCTCAGCGTCAGTTGGT | ||||
| Lactobacillus spp. | Lact-F | CGATGAGTGCTAGGTGTTGGA | 186 | 60 | Fu [96] |
| Lact-R | CAAGATGTCAAGACCTGGTAAG | ||||
| Lactobacillus rhamnosus | Lrha-F | CTTGCATCTTGATTTAATTTTG | 863 | 55 | Alander[97] |
| Lrha-R | CCGTCAATTCCTTTGAGTTT | ||||
| Prevotella spp. | Pre-F | CACRGTAAACGATGGATGCC | 527 | 55 | Matsuki [91] |
| Pre-R | GGTCGGGTTGCAGACC | ||||
| Total bacteria | Uni331-F | TCCTACGGGAGGCAGCAGT | 467 | 59 | Mohammadi [98] |
| Uni797-R | GGACTACCAGGGTATCTAATCCTGTT |
Table 1: 16S rRNA gene-targeting taxon-specific primers used for quantitative PCR.
DNA amplification and detection by quantitative polymerase chain reaction was performed using power SYBR Green PCR master mix (Applied Biosystems, USA) in optical-grade 96-well plates in a total volume of 20 μL and each PCR reaction was run in triplicate. The PCR master mix contained the following ingredients: Magnesium chloride (5.5 mM/L) 4.4 μL; buffer solution (1.5 mM/L)2 μL; dNTP (0.25mM) 0.4 μL; the appropriate primers (200 nmol/L) 0.8 μL; fecal genomic DNA or pure bacteria DNA 2 μL; SybrGreen 0.2 μL; ROX (50 nM)0.04 μL; Tag (1.25 U) 0.1 μL and DNA free water 10.26 μL. Melting curves for gDNA was generated by the Real Time PCR (ABI 7500 FAST; LifeTechnologies GmbH, Darmstadt, Germany) according to the instruction of the manufacturer.
Real time PCR system
PCR initial heat activation 5 min/95°C, 2-step cycling denaturation 10 s/95°C, combined annealing/extension 30 s/60°C and the number of cycles was 35–40.qPCR standard curves were constructed for each primer pair using 10-fold serial dilutions of bacterial genomic DNA of known concentration. The qPCR data were converted to the estimate of log10 total genome copies from each bacterial taxon present in one gram fecal wet weight using the appropriate software program.
Analysis of fecal short chain fatty acids (SCFAs): SCFAs were extracted from fresh feces (300 mg) with six-fold volume of perchloric acidified water in the presence of 23.6 μl isobutyric acid (12 mM) as an internal standard. Following vortexing and lyophilization, SCFAs were extracted with diethylether and silylated. The samples were allowed to stand for 48 h before injection to complete the derivatization. The gas chromatograph (HP 5890 series Hewlett-Packard, Waldbronn, Germany), equipped with a HP-20 M column (25 m × 32 mm; film thickness 0.3 μm) and a flame ionization detector, was used for SCFA measurements. Helium was used as the carrier gas at a flow rate of 1 ml min-1. The initial column temperature was 75°C for 1 min; the temperature was increased to 100°C at a rate of 20°C min-1 and then to 150°C at a rate of 5°C min-1 and then maintained for 3 min at this temperature. The temperature of the injector and the detector was 200°C and the split ratio was 1:10. SCFA concentrations were expressed as mmol g-1 fresh fecal material.
The lower limit of reliable detection of each product was taken as 0.2 mM. Fecal lactate was assayed enzymatically according to the instruction of the manufacturer of the kit (Boehringer Mannheim- Darmstadt).
Statistical analyses
T-tests were used to compare distribution of values between the groups. General linear modeling was used to test the effect of dietary supplementation (control, SS) and sampling day (0 and 21); volunteer identity was regarded as random effect.
Boxplot whiskers depict minimum and maximum values for each group. P values were calculated using two-tailed T-test, statistically significant (at α=0.05 level) differences are denoted with a star. Abbreviations used: Bact - total Bacteroides , Bif - total Bifidobacterium , Blon - B. longum , Bla - Blautia producta , Cbut - Clostridium butyricum , Clep – total C. leptum group, Ent - total Enterobacteriaceae, Ecol - Escherichia coli, Fpra - Faecalibacterium prausnitzii , Lact - total Lactobacillus , Lrha - L. rhamnosus , Pre - total Prevotella , Ace - acetic acid, Pro - propionic acid, But - butyric acid, Val - valeric acid, iVal - isovaleric acid, Lac - lactate. Note that the estimated values for total Bifidobacterium are lower than those for Bifidobacterium longum due to limited genus coverage of the Bifidobacterium-wide primer set (42% genus coverage based on RDP probe match) and lower efficiency of product formation for that primer pair in qPCR (long amplicon).
Principal components analysis was used to assess the overall metabolite and microbiota composition of fecal samples (Figure 1).
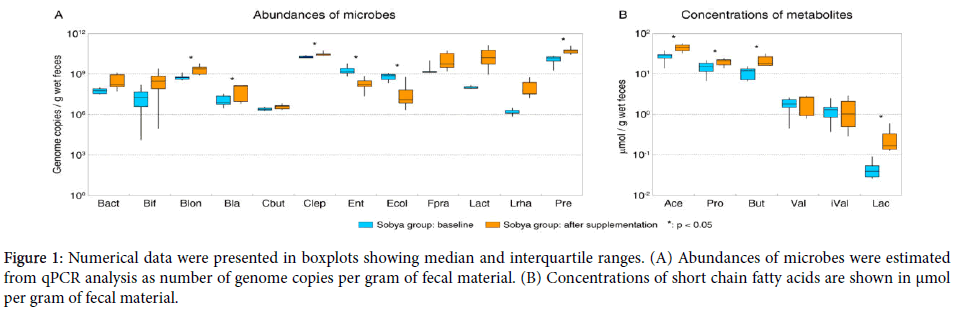
Figure 1. Numerical data were presented in boxplots showing median and interquartile ranges. (A) Abundances of microbes were estimated from qPCR analysis as number of genome copies per gram of fecal material. (B) Concentrations of short chain fatty acids are shown in μmol per gram of fecal material.
Fourteen healthy Egyptian adults were enrolled into a 2-arm diet intervention trial, where one group consumed daily for three weeks fermented SS and the other group served as control.
The two groups of participants were very well matched, being within a narrow age and BMI range, all from the same geographical area and all accustomed to food prepared according to a typical Egyptian cuisine (p>0.05 in all comparisons).
Analysis of the participants’ dietary habits indicated similar consumption patterns of grains, milk products, fruits and vegetables in both groups (data not shown).Quantitative PCR was used to obtain estimates of microbial abundances in all fecal samples at the start of the dietary intervention. Among the profiled taxa, Clostridium leptum group (Clostridium cluster IV, includes Faecalibacterium prausnitzii ), Prevotella , and Enterobacteriaceae taxa were the most abundant (Figure 1A and Table 2). The initial fecal counts of E. coli (8.4 log10 genome copies per g wet feces) were also quite high.
| Phylum | Taxon | Detection | Log10 genome copies / g wet feces | |
|---|---|---|---|---|
| Average ± SD | Median | |||
| Actinobacteria | Bifidobacterium spp. | 14/14' | 7.2 ± 1.7 | 7.3 |
| Bifidobacterium longum | 01-10-2014' | 8.9 ± 0.4 | 8.8 | |
| Bacteroidetes | Bacteroidesspp. | 01-10-2014' | 8.1 ± 0.5 | 7.9 |
| Prevotellaspp. | 14/14' | 9.0 ± 1.1 | 8.9 | |
| Firmicutes | Clostridium butyricum | 14/14' | 6.3 ± 0.1 | 6.3 |
| Clostridium leptum group | 11/14' | 9.9 ± 0.6 | 10.2 | |
| Blautia producta | 14/14' | 7.1 ± 0.4 | 7.2 | |
| Faecalibacterium prausnitzii | 14/14' | 9.4 ± 0.3 | 9.4 | |
| Lactobacillus spp. | 14/14' | 8.3 ± 0.3 | 8.3 | |
| Lactobacillus rhamnosus | 14/14' | 6.4 ± 0.3 | 6.4 | |
| Proteobacteria | Enterobacteriaceae | 13/14' | 9.4 ± 0.3 | 9.4 |
| Escherichia coli | 14/14' | 8.4 ± 0.6 | 8.5 | |
Table 2: Gut microbiota composition at baseline in healthy Egyptian adults.
Microbial composition of SS
The 170 g daily portion of SS provided 4.4 ± 0.62 billion cfu lactic acid bacteria (mostly Lactobacillus rhamnosus ) and 1.79 ± 0.14 billion cfu of yeast. The proteomic spectra of MALDI-TOF identified 5 strains out of 7 LAB isolates as Lactobacillus rhamnosus with high score values. The API analysis identified the sobya yeast as Sachharomy cescerevisia .
Abundances of several groups changed among the participants consuming SS for three weeks compared to the respective control group. In the participants consuming SS, L. rhamnosus transited through the gastrointestinal tract and colonized the intestinal community resulting in an average 46-fold increase in its abundance; no increase in average cell numbers for this species was observed for control group (Table 3).
| Bacterial taxon | Participant group | Log10 genome copies/g wet fecesa | GLMb | |||
|---|---|---|---|---|---|---|
| Day 0 | Day 21 | Group | Day | Diet x Day | ||
| Bacteroides spp. | CONT | 8.5 | 8.5 | - | - | - |
| SS | 7.7 | 8.4 | ||||
| Bifidobacterium spp. | CONT | 7.8 | 8.7 | - | - | - |
| SS | 6.6 | 8 | ||||
| Bifidobacterium longum | CONT | 9.1 | 9.1 | - | 0.01 | - |
| SS | 8.7 | 9.3 | ||||
| Blautiaproducta | CONT | 7.1 | 7.3 | - | 0.01 | - |
| SS | 7 | 7.6 | ||||
| Clostridium butyricum | CONT | 6.3 | 6.4 | 0.01 | 0.01 | - |
| SS | 6.4 | 6.6 | ||||
| Clostridium leptum group | CONT | 9.2 | 9.7 | 0.01 | - | - |
| SS | 10.3 | 10.5 | ||||
| Enterobacteriaceae | CONT | 9.5 | 8 | - | 0.01 | - |
| SS | 9.2 | 8.2 | ||||
| Escherichia coli | CONT | 8 | 7.9 | 0.03 | 0.01 | 0.01 |
| SS | 8.8 | 7.4 | ||||
| Faecalibacterium prausnitzii | CONT | 9.5 | 9.7 | - | 0.04 | - |
| SS | 9.3 | 10 | ||||
| Lactobacillus spp. | CONT | 8.5 | 8.7 | - | - | - |
| SS | 8 | 10.1 | ||||
| Lactobacillus rhamnosus | CONT | 6.6 | 6.6 | - | - | - |
| SS | 6.2 | 7.8 | ||||
| Prevotella spp. | CONT | 8 | 8.9 | 0.01 | 0.02 | 0.02 |
| SS | 10 | 10.7 | ||||
Table 3: Mean initial and final bacterial taxon abundances among the control and SS groups (a: Mean among seven subjects; b: Results of multivariate general linear modeling (GLM) with group and day used as fixed factors; “-“ – not significant at α=0.05 level).
This finding is in line with reports from Finish intervention study, which found that participants excreted on average more than 1000-fold higher numbers of L. rhamnosus GG at the end of a three week dietary supplementation with 1010 cfu of L. rhamnosus GG compared to the placebo group [Lahti]. In addition to lactic acid bacteria increase after the intake of SS, the counts of Bifidobacterium longum , Blautia spp., Clostridium leptum , Faecalibacterium prausnitzii , and Prevotella spp. also increased significantly (p<0.05, Figure 1A). Interestingly, fecal Faecalibacterium prausnitzii could not be detected in two subjects assigned to SS group at day 0; however, F. prausnitzii was restored after the three-week intervention with SS. SS intake was also associated with significant reduction (p<0.05) in the fecal counts of total Enterobacteriaceae and specifically E. coli compared with respective baseline counts (Figure 1A).
Concentrations of short chain fatty acids
Levels of three major SCFAs, butyrate, propionate, and acetate, all increased after consumption of SS (p<0.05, see Figure 1B). No respective changes were observed among the control group. Expectedly, levels of lactate were also higher after SS administration, though the overall concentration of lactate in stool was low due to likely reuse of this fermentation product by secondary and tertiary degraders [65]. Initial fecal pH values averaged 5.9 and were reduced to 5.3 (p<0.05) at the end of the three week dietary intervention; the respective fecal pH values for the control group remained almost unchanged.
Ordination analysis of microbial and metabolite datasets
The microbial abundances and metabolite concentration values among all samples were subjected to principal components analysis to assess the similarity of microbial and metabolite profiles among samples [66].
Figure 2A shows the PCA output for microbial dataset, and Figure 2B visualizes metabolite-based PCA. Samples collected from sobya group after SS supplementation displayed larger shift in their microbial and metabolite profiles compared to the control group. To assess this observation statistically, we calculated for each sample the difference of its PC1-PC2 coordinates (=distance in PCA space) at day 0 and day 21, and show the distribution of these distances for sobya and control groups in Figure 2C. This analysis confirmed that sobya group samples were separated significantly more in the PCA space than those from the control group.
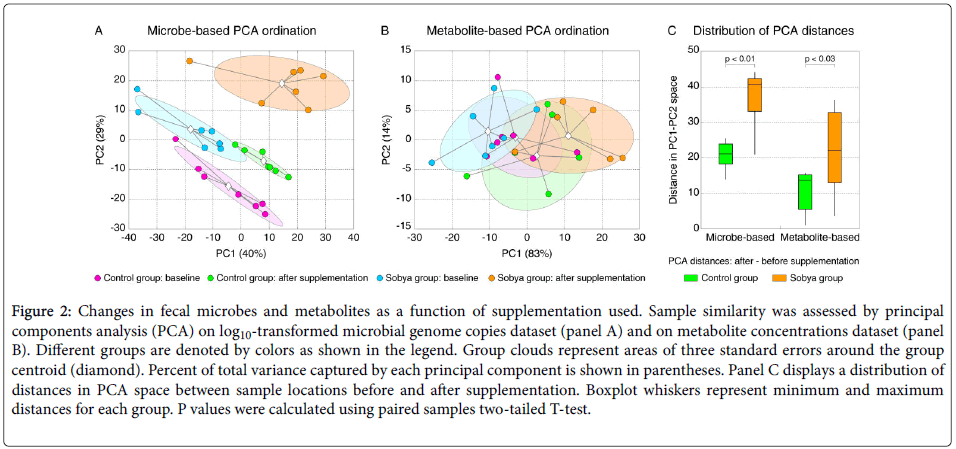
Figure 2. Changes in fecal microbes and metabolites as a function of supplementation used. Sample similarity was assessed by principal components analysis (PCA) on log10-transformed microbial genome copies dataset (panel A) and on metabolite concentrations dataset (panel B). Different groups are denoted by colors as shown in the legend. Group clouds represent areas of three standard errors around the group centroid (diamond). Percent of total variance captured by each principal component is shown in parentheses. Panel C displays a distribution of distances in PCA space between sample locations before and after supplementation. Boxplot whiskers represent minimum and maximum distances for each group. P values were calculated using paired samples two-tailed T-test.
The present study explored the fecal microbiome diversity of healthy Egyptian adults and tested the impact of bacterial quantification based on real-time PCR provides a challenging analytical strategy for monitoring the microbial composition in response to dietary interventions. The application of PCR since the early 1990s in the combination with the extraction of genomic DNA from fecal specimens has been central to the development of culture independent approaches in microbial ecology [67]. In the present study, the fluorescence-based Q-RT-PCR was utilized to record the increase in the fluorescent signal associated with amplicons formation during each cycle of the PCR amplification and hence facilitating quantitative determination of the initial template gene numbers. Quantification of the target gene during exponential amplification avoids problems that are associated with so-called ‘end-point’ PCR. The minimum information required for Q-RT-PCR (MIQE) that includes a set of guidelines in the form of checklist for evaluating qPCR experiments [68] were followed and the validity of the protocols was proven. The selection of primer pairs was based on previously published data and was tested in the present study against all reference bacteria listed in the materials and methods section by using optimized reaction conditions for each assay and subsequent melt curve analysis. The assays were positive for the corresponding target species and no crossreactions were observed with any of the nontarget micro-organisms of ca. 1,600 bp available in GenBank release 185.0 [69].
The abundance of fecal Prevotella group was high in all participants, in line with the fact that gut Prevotella correlates positively with a Mediterranean diet, specifically with high levels of carbohydrate, fruit, and vegetable intake [70]. While fecal Bacteroides species were detected in most participants, their numbers were much lower, consistent with the fact that Bacteroides are abundant among populations consuming animal-based diet [71]. The two genera of the Bacteroidetes: Bacteroides and Prevotella were reported to be antagonistic to each other [72], and our findings support that conclusion. The present results are also consistent with the previous report on the differences in gut microbiota of American and Egyptian adolescents [73] and the high abundances of enteric bacteria in the fecal samples among Egyptians indicating potential higher load of pathogenic gut microorganisms .The high counts of fecal Lactobacillus and its ratio to the total anaerobic bacteria are typically used as an indicator of healthy gut [74]. Lactobacillus rhamnosus GG (ATCC 53103) is one of the most thoroughly studied probiotics and its beneficial effects include prevention of antibiotic-associated diarrhea, treatment and prevention of rotavirus diarrhea and enhancement of intestinal immunity. The ability of strain GG to survive passage through the gastrointestinal tract has been demonstrated in both adults and children by the use of fecal samples [73]. The intake of Lactobacillus acidophilus improved the intestinal permeability after daily intake for 75 days by Egyptian children [53].
The growing popularity of fermented cereal products and considerable health and food safety trends in recent years have emphasized the importance of knowing which food - type microbes are involved in the production of SS. For the enumeration of Lactobaccilli in fermented cereals, lactic acid bacteria (LAB) was enumerated on de Man, Rogosa and Sharp (MRS) agar media without [46], or in the presence of 0.1 g/ l [75], 0.2 g/ l [76] or 5 g/L [77] filtersterilized cycloheximide to inhibit yeast growth . In the present study, LAB was grown on MRS agar plates with the addition of 3 % ethanol and 0.5 % cycloheximide [59,77]. The incorporation of cycloheximide in the MRS media increased the Lactobacilli count by 2.3 fold excess (1.56 ± 0.22 and 4.4 ± 0.62 billion cfu per 170 g portion size in the absence and presence of 0.5 % cycloheximide and 3% ethanol, respectively).
The microbial composition of sobya shows population variations depending on the origin of grains; wheat in Saudi Arabia [45] and rice in Egypt [51]. A symbiotic relationship exists between the bacteria and the yeasts and their combined growth results in the fermented sour sobya (SS) with distinct flavour characteristics and acidic pH of 3.5. Consumption of SS promoted higher abundance of Lactobacillus spp., Bifidobacterium longum , and Faecalibacterium prausnitzii human gut microbiota members associated with improved health [78] and drastically reduced the numbers of total Enterobacteriaceae by 12-fold and E. coli by 26-fold. Other beneficial effects of sobya supplementation included significant increase in fecal butyrate concentration relative to the respective baseline levels. Butyrate is instrumental in mucosal integrity [16] and is the principal source of metabolic energy for the colonocytes. It modulates gut homeostasis [80], promotes removal of dysfunctional cells [16], promotes genomic stability and potentially protects against colon cancer [81]. Since lactobacilli present in the supplemented SS are not butyrate producers, we speculate that LAB stimulates SCFA production through increased cross-feeding with butyrate producers belonging to the Clostridium cluster IV [82]. Alternatively, certain butyrate producing taxa could be favored by the lowering in colonic pH values following the dietary intervention with SS [83].
Today, there is a trend for changing the probiotics from dairy-based products to whole grain- based functional foods [84]. L. rhamnosus isolated from SS are characterized with robust sustainability [85,86] and may open exciting perspectives for industry-driven applications in wide range of fermented food matrices. The ultimate goal is to use microbiome-based therapies in nutrition [87]. Yoba 2012 was reported to be the world’s first generic probiotic strain [88] formulated by culturing L. rhamnosus GG with Streptococcus thermophilus to optimize synergistic propagation of both strains in wide range of fermented foods [89].
The unique characteristics of fermented SS used in the present study with its richness in synergistic effects between positively alter host metabolism and gene expression and might be useful as an adjunctive therapy in addition to other nonpharmacological interventions for the management of gastrointestinal disorders, mild malnutrition and high oxidative burden in childhood. The promotion of a healthy lifestyle by non-pharmacological means is a preventive measure strategy aiming at good health.
We are thankful to all volunteers for collaboration during participation in the dietary intervention trial. We are very grateful to Dr. Wilhelm Bockelmann from the Max Rubner Institute for Microbiology and Biotechnology Kiel-Germany for the identification of the sobya yeast. Thank is extended to Dr. Mohmoud mohamad for the supply of the cycloheximide and to Dr Hoda Mabrok from the Nutrition Department, NRC for revising the sequence of the primers .
Citation: Labib E, Blaut M, Hussein L, Gouda M, Kramer DL, et al. (2018) Molecular Diversity of Gut Microbiota and Short Chain Fatty Acids in Egyptian Adults Following Dietary Intervention with Fermented Sobya. J Food Microbiol Saf Hyg 3: 139.
Received: 06-Apr-2018 Accepted: 27-Sep-2018 Published: 01-Oct-2018
Copyright: © 2018 Labib E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.