
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research - (2019)Volume 5, Issue 2
Aim: The complexity of multifactorial diseases, such as cancer, poses significant challenges to the development of personalised therapies. Integrated digital histological analysis of tumours provides a better understanding of the immune microenvironment and the prognostic relationship associated with the enumeration and distribution of specific tumour infiltrating lymphocyte (TILs) subpopulations. To this effect multiplex cell labelling , alongside multi-spectral imaging (MSI) is an approach increasingly used to achieve more accurate in-situ TIL phenotyping and quantification. However, these approaches require full validation prior to utilisation , which is the fundamental aim of this study.
Methods: Whole sections and tissue microarrays of lymphocyte-rich tissue were used to develop and validate a multiplex immunofluorescence (IF) protocol for simultaneous MSI interrogation of up to six immune cell antigens of interest; CD3, CD8, FOXP3, CD20, PD-L1 and PD1. Concordance between single- plex chromogenic immunohistochemistry (IHC) and single- plex immunofluorescence (IF) staining was first achieved. Subsequently, the effect of the position in a multiplex IF order for any given antibody was investigated, understanding the impact of antibody steric hindrance and antibody stripping conditions.
Results: In methods where multiplexing is enabled using antigen retrieval to strip prior antibodies, the principal compounding factor influencing multiplex-assay validation was the non-linear and non-uniform effect of extended times of heat-induced epitope retrieval (HIER), as antibodies advance in order in a multiplex protocol.
Conclusion: This study demonstrates the fidelity of multiplex staining as representative of single- plex staining, the effect of order of antibody staining, and offers a framework for the generation of optimised multiplex immunofluorescent protocols.
Multiplex immunofluorescence; Digital histology; Alidation; Immune infiltrates
The Tumour Microenvironment (TME) is important in tumour progression and treatment response, leading to development of new targeted therapies [1,2]. Tumour infiltrating lymphocytes (TILs) are a common feature of solid cancers with their type, number and spatial distribution all shown to affect prognosis [3,4]. Similarly, clinically validated quantification of CD3+ lymphocytes and CD8+ cytotoxic T cells has shown statistical superiority to the current TNM classification for prediction of overall survival (OS) and disease-free survival (DFS) in colorectal cancer [1,5,6]. However, to gain a deeper understanding of the TME, further characterisation is needed, including identification of phenotypically distinct immune cell populations such as dendritic cells, macrophages, natural killer cells, and their state of activation or exhaustion. The ability to simultaneously assess multiple cells in situ is dependent on development of accurate, sensitive and quantifiable multiplexing assays.
Multiplex immunostaining, whether chromogenic or fluorescent, is becoming increasingly popular with the emergence of robust multiplex staining methods and advanced multispectral imaging [7-9]. Multiplex staining has significant advantages in terms of preservation of precious tissue samples, and co-localisation of antigens [10]. The latter is of particular importance for the growing field of immunotherapy, enabling detection and identification of tissue infiltrating cells, along with characterisation of response biomarkers such as PD-L1 [8,10-13]. However, stringent validation steps must be implemented when developing multiplex assays to ensure these assays generate comparable data to the gold-standard single-plex assays.
This project describes development and validation of a multiplex staining protocol against CD3, CD8, FOXP3, CD20, PD-L1 and PD1, using a multiplex methodology that includes sequential rounds of antibody stripping. Critically it highlights the challenges of developing multiplex assays and stresses the importance of not using a ‘plug and play’ approach, which can result in inaccurate data read-outs. Measurement of the fidelity of multiplex staining as representative of single-plex staining, the effect of antigen retrieval time and the order of antibody staining are critical to the generation of a robust optimised multiplex immunofluorescence (IF) protocol.
Tissue cohorts and ethics
Cohort 1: Whole tissue sections of tonsil. Ethical approval was provided by the Central Manchester Multicentre Research Ethical Committee (03/TG/076).
Cohort 2: A follicular-lymphoma tissue microarray (TMA) constructed from 40 archived pre-diagnostic FFPE biopsies (82 cores). Ethical permission for this study was obtained from the Central Manchester Multi-Centre Research Ethical Committee (03/8/106).
Control tissue: Samples were acquired via the AstraZeneca Biobank [Human Tissue Authority (Licence No. 12109) and National Research Ethics Service Committee (NREC) Approval as a Research Tissue Bank (RTB) (REC No 17/NW/0207)].
Chromogenic single-plex immunohistochemistry staining
Chromogenic staining (DAB single-plex) was performed on the Ventana Discovery Ultra autostainer (Roche) using UltraView DAB (Roche) or Chromomap DAB (Roche) detection kits. 4 μm whole tissue FFPE sections were mounted on SuperFrost™Plus slides and dried over-night at 37°C prior to staining. The Ventana staining protocol involved EZ prep deparaffinisation, followed by CC1 antigen retrieval (pH 9, 95°C), then endogenous peroxidase blocking (Roche), followed by incubation with the primary antibodies under the conditions listed in Table 1 and then chromogenic detection following the manufacturer’s recommendation for each kit. Counterstaining was performed using Hematoxylin II (Roche) and then bluing reagent (Roche).
| Antibody | Species | Clone | Manufacturer | Incubation time (mins) |
|---|---|---|---|---|
| CD3 | Rabbit | 2GV6 | Roche | 16 |
| CD8 | Mouse | C8/144b | Dako | 32 |
| CD20 | Mouse | L26 | Roche | 16 |
| PD1 | Mouse | NAT105 | Abcam | 60 |
| PDL1 | Rabbit | SP263 | Roche | 16 |
| FOXP3 | Mouse | 236A/E7 | Abcam | 60 |
Table 1: List of primary antibodies used, and their incubation details.
Staining of single-plex, duplex and multiplex immunofluorescence
IF staining was performed on the Ventana Discovery Ultra autostainer, with the 6-plex achieved using three sequential dualplex protocols. Tissues were sectioned, deparaffinised, antigen retrieved and blocked for endogenous peroxidase as described above. The primary antibodies used, and their incubation details, are listed in Table 1.
Following incubation with the first primary antibody, slides were incubated with a horseradish peroxidase labelled secondary antibody, followed by disclosure using tyramide-linked fluorescent marker. For duplex and multiplex staining this was followed by heat-induced epitope retrieval (HIER) prior to addition of the second primary antibody, followed by incubation with a horseradish peroxidase labelled secondary antibody and finally disclosure using the next tyramide-linked fluorescent marker. Fluorescent detection was performed using Opal 7-plex fluorophores (PerkinElmer, Waltham, Massachusetts, USA). Opal fluorophores were supplied premixed with dimethyl sulfoxide (DMSO) and were diluted 1:75 with tyramide signal amplification fluid (TSA). Details of the fluorophores used and the primary antibodies they were designated to for the 6-plex stain, are shown in Table 2 (note, different fluorophore-antibody matches were used in the single-plex and duplex stains). For the 6-plex stain, following secondary antibody and fluorescent marker detection of the sixth antibody, sections were mounted with ProLong Gold Antifade Mountant with DAPI (Molecular MolProbes, Eugene, Oregon, US) and cover slipped. For the single-plex and duplex stains, DAPI (PerkinElmer) was applied manually before mounting with ProLong Diamond Antifade Mountant (ThermoFisher, Waltham, Massachusetts, US) and cover slip addition.
| Fluorophore | Excitation wavelength (nm) | Emission wavelength (nm) | Designated antigen |
|---|---|---|---|
| Opal520 | 494 | 525 | CD3 |
| Opal540 | 523 | 536 | CD8 |
| Opal570 | 550 | 570 | FOXP3 |
| Opal620 | 588 | 616 | CD20 |
| Opal650 | 627 | 650 | PD1 |
| Opal690 | 676 | 694 | PD-L1 |
Table 2: Details of the fluorophores used and the primary antibodies they were designated to for the 6-plex stain.
HALO image analysis of single-plex chromogenic immunohistochemistry to determine HIER condition
The single-plex slides for each antibody at each HIER condition were digitalised using an Aperio AT2 whole slide scanner (Leica) at 20x objective. The HALO® image analysis platform was used to quantitatively analyse the protein expression, with stains classified as either cyto-nuclear, membrane or immune. Consecutive tissue sections were co-registered using a rigid-body registration method within the HALO platform and protein expression measured using colour deconvolution to separate DAB and Hematoxylin staining based on optical density. The number of cells positive for each antibody was compared between different durations of HIER with linear regression analysis using Graphpad Prism version 6.0 (Graphpad Software Inc, San Diego, California, USA).
HALO Image analysis of single-plex and duplex immunofluorescence
Single-plex and duplex IF stained slides were digitalised using the AxioScan Z1 (Zeiss). HALO® image analysis platform in combination with the cytonuclear fluorescence algorithm was used to analyse protein expression of each fluorescent stain across serial tissue sections. The number of cells positive for each antibody was compared between different durations of HIER with linear regression analysis using Graphpad Prism version 6.0.
Inform image analysis of multiplex immunofluorescence
Multispectral image analysis was performed using Inform software version 2.1.1 (PerkinElmer, Waltham, Massachusetts, US). Cell-based segmentation was used with nuclear segmentation, based on DAPI counterstaining. For Cohort 1 (whole sections), five regions of interest (ROI) were selected from each DAB stained section and were matched across singleplex fluorescent and multiplex stained sections (Figure 2b). For Cohort 2 the entire TMA was imaged at x200 magnification using a Vectra multispectral microscope and after cell phenotyping and spectral un-mixing, the total number of positive cells for each antibody were counted for each core and divided by the area of tissue in the core. The mean cell counts per tissue area were then compared for each antibody with those for the same antibody applied to the same TMA in single-plex IF. A spectral library composed of the spectra of each of the six Opal fluorophores singly was generated, which could then be used together with spectra of DAPI and tissue auto fluorescence. This was applied with spectral un-mixing to generate positivity, followed by phenotype training to identify cells positive for each antibody. The trained Inform protocol was applied to the five ROI and the number of positive cells for each antibody was calculated between the single-plex and multiplex stained sections.
Statistical analysis
The strength of association between the numbers of cells positive for each antibody as indicated by single-plex and multiplex sections was assessed by linear regression (only for 6- plex experiment) analysis using Graphpad Prism version 6.0. R2 values from regression analysis was used for the final 5 plex comparisons. Coefficient of variability was determined using Graphpad Software for duplex IF and single-plex serial section IF experiments.
Investigation of effect of antibody order in multiplex fluorescent immunofluorescence
Prior to considering validation of a full multiplex IF protocol; it was first essential to demonstrate concordance between singleplex chromogenic (DAB) IHC and single-plex IF staining. Quantification of CD3, CD8 and FOXP3 on control tonsil tissue demonstrated concordance between the two staining protocols, which fell within expected intra-run variability (Figures 1A-1F). Subsequently, the effect of the position in a multiplex IF order for any given antibody was investigated across a panel of six antibodies, selected to detect a range of nuclear and membrane markers characteristic of infiltrating immune cells. Specifically, these were CD3+ lymphocytes, CD20+ B cells, CD8+ cytotoxic T cells, Foxp3+ regulatory T cells (Tregs), PD1 (program death 1) and PD-L1 (program death-ligand 1).
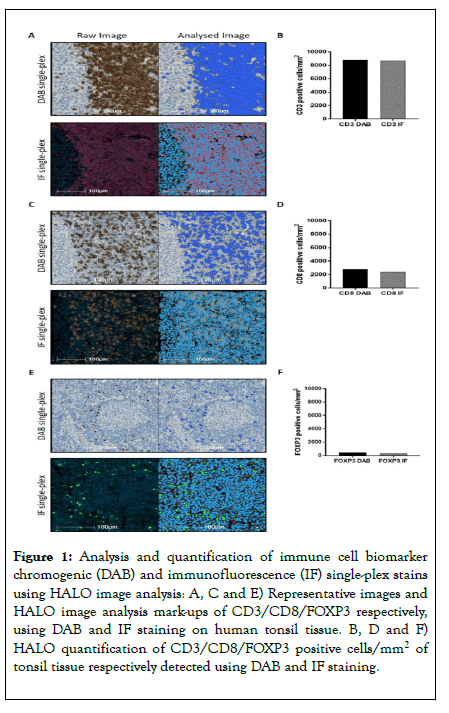
Figure 1. Analysis and quantification of immune cell biomarker chromogenic (DAB) and immunofluorescence (IF) single-plex stains using HALO image analysis: A, C and E) Representative images and HALO image analysis mark-ups of CD3/CD8/FOXP3 respectively, using DAB and IF staining on human tonsil tissue. B, D and F) HALO quantification of CD3/CD8/FOXP3 positive cells/mm2 of tonsil tissue respectively detected using DAB and IF staining.
For each position in the multiplex staining order (1 to 6), a comparison was made on serial sections, between the changing multiplex position and single-plex IF (Table 3). Five regions with variable immune cell densities were selected from whole tissue sections of cohort 1, aligned across the slides being compared. The single-plex methodologies were applied in this multiplex system without further protocol optimisation. Unfortunately, this resulted in PD-L1 run fails and this antibody was therefore omitted from analysis, leaving data for five of the six experimental runs. For CD3, CD8, CD20, FOXP3 and PD1 the R2 values from regression analysis presented a trend to progressive reduction as the respective antibody advanced in position (Figures 2A-2E). This indicates a reduction in fidelity of antibody staining as the antibody was applied progressively later in the multiplex staining order.
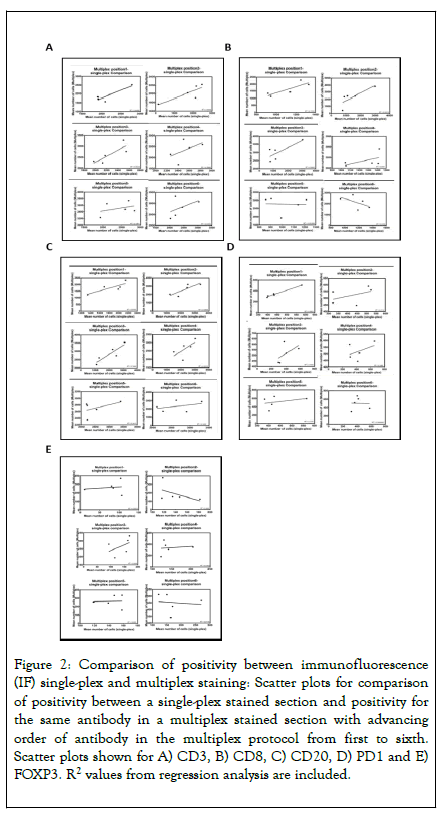
Figure 2. Comparison of positivity between immunofluorescence (IF) single-plex and multiplex staining: Scatter plots for comparison of positivity between a single-plex stained section and positivity for the same antibody in a multiplex stained section with advancing order of antibody in the multiplex protocol from first to sixth. Scatter plots shown for A) CD3, B) CD8, C) CD20, D) PD1 and E) FOXP3. R2 values from regression analysis are included.
| R2 figures for multiplex IF vs. Single-plex IF at the following positions | ||||||
|---|---|---|---|---|---|---|
| Antibody | Position 1 | Position 2 | Position 3 | Position 4 | Position 5 | Position 6 |
| CD3 | 0.807 | 0.7 | 0.683 | 0.267 | 0.136 | 0.241 |
| CD8 | 0.74 | 0.651 | 0.596 | 0.301 | 0.002 | 0.313 |
| CD20 | 0.679 | 0.697 | 0.789 | 0.431 | 0.242 | 0.14 |
| FOXP3 | 0.021 | 0.249 | 0.203 | 0.016 | 0.002 | 0.012 |
| PD1 | 0.98 | 0.212 | 0.309 | 0.196 | 0.115 | 0.001 |
| PD-L1 | NA | NA | NA | NA | NA | NA |
Table 3: Comparison on serial sections, between the changing multiplex position and single-plex IF.
Investigation of biological/run variability in serial sectioning, antibody steric hindrance and HIER conditions in duplex IF
Several experimental hypotheses were tested to help understand the reasons behind the variability in antibody staining observed in Figure 2. Through combination of membrane markers (CD3 or CD8) with nuclear FOXP3 in duplex staining on control tonsil tissue, we confirmed variability from staining order was not simply attributable to biological heterogeneity between serial sections. For both CD3 and CD8 assays, variability of four serial sections stained with respective single-plex DAB, single-plex IF and duplex IF (membrane marker at position 1 or 2) was compared with single-plex IF variability across three serial sections. The coefficient of variability (CV) for single-plex IF serial sections was comparable to duplex assay CV for CD3 (Figures 3A-3C) but not CD8 (Figures 3D-3F). Of note, detection of CD8 when placed in position 2 saw a 1.6 fold increase from single-plex IF quantification. This again demonstrates the importance of staining order in a multiplex assay, where in this case, CD8 must be stained in position 1 to replicate single-plex quantification.

Figure 3. HALO image analysis was used to assess the impact of staining order and steric hindrance in immunofluorescence (IF) duplex stains: A) Representative image and HALO image analysis mark-up of CD3/FOXP3 IF duplex staining on human tonsil tissue, where CD3 (red) was stained in position 1 and FOXP3 (green) was in position 2. B) HALO quantification of CD3 positive cells/mm2 tissue in chromogenic (DAB) CD3 single-plex, IF CD3 single-plex and IF duplex stains where CD3 was placed in either position 1 or 2 in combination with FOXP3. Percentage coefficient of variation (CV) was calculated across the samples. C) Intra-run variability of CD3 IF stained serial tonsil sections; CD3 positive cells/mm2 tissue and the CV are shown. D) Representative image and HALO image analysis mark-up of CD8/FOXP3 IF duplex staining on human tonsil tissue, where CD8 (orange) was stained in position 1 and FOXP3 (green) was in position 2. E) HALO quantification of CD8 positive cells/mm2 in DAB CD8 single-plex, IF CD8 single-plex and IF duplex stains where CD8 was placed in either position 1 or 2 in combination with FOXP3. CV also shown. F) Intra-run variability of the CD8 single-plex IF staining of three serial tonsil sections stained for CD8 using IF (CV also shown). G) Representative image and HALO image analysis mark-up of CD3/CD8 IF duplex staining on human tonsil tissue, where CD3 (orange) was stained in position 1 and CD8 (green) was in position 2. H) HALO quantification of CD3 positive cells/mm2 tissue in DAB CD3 single-plex, IF CD3 single-plex and IF duplex stains where CD3 was placed in either position 1 or 2 in combination with CD8. I) Representative image and HALO image analysis mark-up of CD8/CD3 IF duplex staining on human tonsil tissue, where CD8 (green) was stained in position 1 and CD3 (orange) was in position 2. J) HALO quantification of CD8 positive cells/mm2 tissue in DAB CD3 single-plex, IF CD3 single-plex and IF duplex stains where CD8 was placed in either position 1 or 2 in combination with CD3.
Added to this, application of antibodies that target the same cells and sub-cellular compartment are thought to cause steric hindrance due to the proximity of the antibody binding, a potential problem for multiplexing membrane markers such as CD3 and CD8 which have biological overlap of the antigen. Staining and quantification of CD3 when duplexed with CD8 (Figures 3G and 3H) showed minimal variation at either position 1 or 2 compared with single-plex CD3 DAB or IF (CV 6.99%). In contrast, when quantifying CD8 in duplex with CD3 (Figures 3I and 3J), a 90% drop in CD8 positive cells was detected when CD8 was placed at position 2 after CD3. This resulted in a high CV across the experimental groups of 59.51%, which was not observed when CD8 was combined with nuclear FOXP3 (Figure 3E), and suggests steric hindrance is a problem for CD8 when combined with CD3.
An additional consideration was the effect of the increased HIER needed to strip previously stained antibodies. Serial sections were stained for each antibody as a single-plex DAB assay, subjected to HIER durations corresponding to the exposure times at positions 1 through to 6 (increments of 8 minutes per position) and cells quantified using image analysis (Figure 4). The effect of HIER duration was non-linear and nonuniform across the range of antibodies, with all the antibodies showing both a rise and fall in immune cell counts as duration of HIER increased. Of note, the DAB clinical protocol for CD20 doesn’t require HIER and subsequently when HIER was included, the staining specificity was completely lost (Figure 4C). Due to this, CD20 was removed from this multiplex protocol and classed as incompatible to multiplex. The HIER data are summarized in Figure 5.
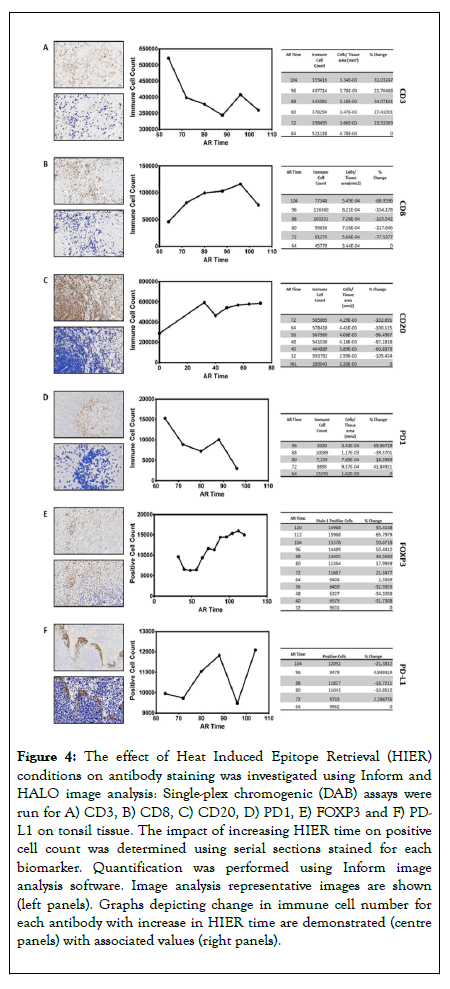
Figure 4. The effect of Heat Induced Epitope Retrieval (HIER) conditions on antibody staining was investigated using Inform and HALO image analysis: Single-plex chromogenic (DAB) assays were run for A) CD3, B) CD8, C) CD20, D) PD1, E) FOXP3 and F) PDL1 on tonsil tissue. The impact of increasing HIER time on positive cell count was determined using serial sections stained for each biomarker. Quantification was performed using Inform image analysis software. Image analysis representative images are shown (left panels). Graphs depicting change in immune cell number for each antibody with increase in HIER time are demonstrated (centre panels) with associated values (right panels).
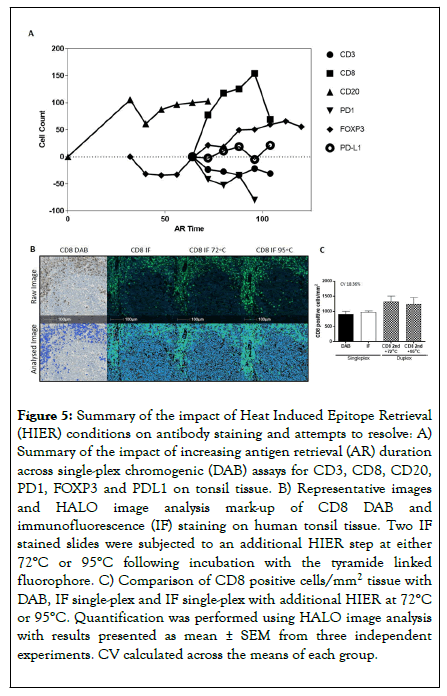
Figure 5. Summary of the impact of Heat Induced Epitope Retrieval (HIER) conditions on antibody staining and attempts to resolve: A) Summary of the impact of increasing antigen retrieval (AR) duration across single-plex chromogenic (DAB) assays for CD3, CD8, CD20, PD1, FOXP3 and PDL1 on tonsil tissue. B) Representative images and HALO image analysis mark-up of CD8 DAB and immunofluorescence (IF) staining on human tonsil tissue. Two IF stained slides were subjected to an additional HIER step at either 72°C or 95°C following incubation with the tyramide linked fluorophore. C) Comparison of CD8 positive cells/mm2 tissue with DAB, IF single-plex and IF single-plex with additional HIER at 72°C or 95°C. Quantification was performed using HALO image analysis with results presented as mean ± SEM from three independent experiments. CV calculated across the means of each group.
A subsequent assay was developed to test the temperature required to achieve successful stripping, and if this could be reduced to limit antibody impact. The cut-off for successful removal of antibody conjugates was determined to be 72°C (data not shown). However, analysis of serial sections stained for CD8 as single-plex DAB, single-plex IF, or single-plex IF with an additional round of stripping revealed no statistical significance in CD8 detection using 72°C rather than routinely used 95°C (Figure 5C). Reducing stripping temperature therefore did not rescue the over-retrieval effect of staining position on CD8, with higher CD8 levels quantified in the multi-plex compared to the gold standard single-plex assays.
Building on the aforementioned data, a multiplex order was suggested of CD8, PD-L1, CD3, FOXP3 and PD1. The multiplex IF protocol detailed above was repeated with the antibodies applied in this order using a tissue microarray composed of 100 cores of lymphoid tissue containing follicular lymphoma (Cohort 2). Single-plex DAB compared to single-plex IF resulted in R2 correlations ranging from 0.19 to 0.72 (Figure 6 and Table 4). When comparing single-plex IF to multiplex IF the R2 correlations ranged from 0.21 to 0.81, demonstrating a good correlation using the optimised protocol. Representative images of the optimised protocol staining are illustrated in Figure 7, showing single DAB, single-plex IF, and multiplex IF and an associated IF artificial bright field image for each marker.
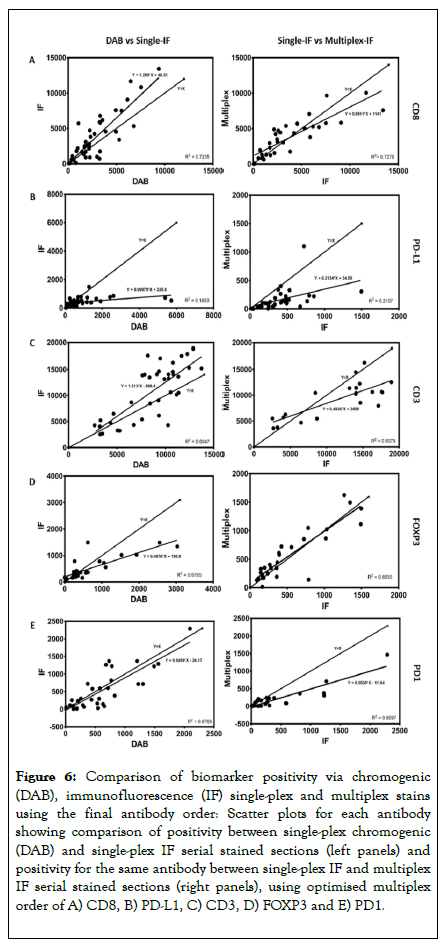
Figure 6. Comparison of biomarker positivity via chromogenic (DAB), immunofluorescence (IF) single-plex and multiplex stains using the final antibody order: Scatter plots for each antibody showing comparison of positivity between single-plex chromogenic (DAB) and single-plex IF serial stained sections (left panels) and positivity for the same antibody between single-plex IF and multiplex IF serial stained sections (right panels), using optimised multiplex order of A) CD8, B) PD-L1, C) CD3, D) FOXP3 and E) PD1.
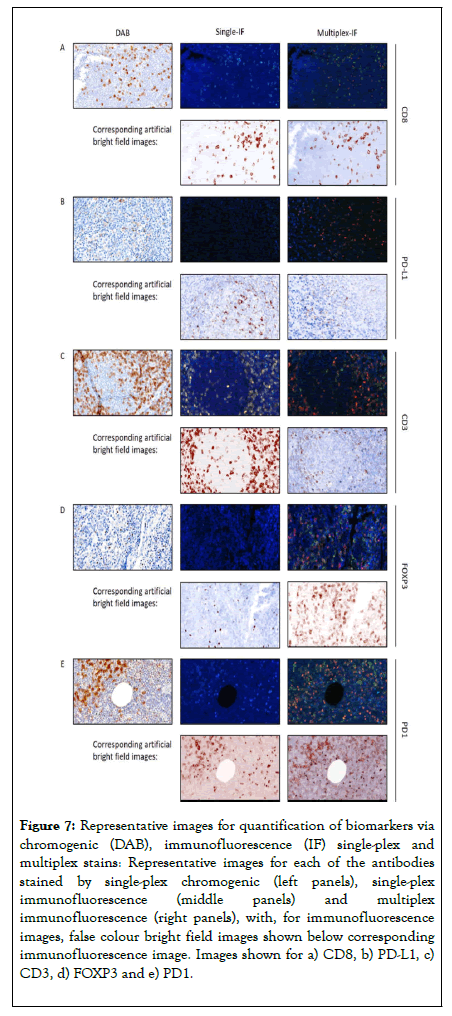
Figure 7. Representative images for quantification of biomarkers via chromogenic (DAB), immunofluorescence (IF) single-plex and multiplex stains: Representative images for each of the antibodies stained by single-plex chromogenic (left panels), single-plex immunofluorescence (middle panels) and multiplex immunofluorescence (right panels), with, for immunofluorescence images, false colour bright field images shown below corresponding immunofluorescence image. Images shown for a) CD8, b) PD-L1, c) CD3, d) FOXP3 and e) PD1.
Multiplex IF staining is increasingly being used for simultaneous analysis of multiple biomarkers [9,14]. It is particularly useful for investigation of TILS, which by their nature require colocalisation of multiple markers for their identification, making multiplex IF essential for their detection [15-17]. Previous studies have evaluated whether the staining for any given antibody in a multiplex protocol equates qualitatively to that when used singly [18], but only recently have studies advanced into understanding whether multiplex staining recapitulates single-plex staining quantitatively or the impacting of antibody stripping [19-22]. In the present study we quantitatively test the effect of antibody order in multiplex staining, showing that it is not feasible to simply merge clinically validated single-plex DAB protocols into a multiplex methodology. As such, thorough re-optimisation and validation will be required to ensure the accuracy and sensitivity of any multiplex assays.
This study indicates, in methods where multiplexing is enabled using antigen retrieval to strip prior antibodies, that the principal compounding factor influencing multiplex validation is the effect of extended times of HIER as antibodies advance in order in a multiplex protocol. Sequential HIER steps occur between each antibody and duration of HIER had a qualitative effect on immune cell counts for all antibodies, the effect being non-linear and non-uniform, with all the antibodies showing both rise and fall in immune cell counts as duration of HIER increased. A reduction of the HIER temperature did not resolve the impact on susceptible biomarkers such as CD8. These data enabled design of an optimised antibody order for the multiplex IF protocol, with minimisation of the effect of increasing duration of HIER by placing antibodies more affected earlier in the multiplex order.
The final optimised antibody order was CD8, PD-L1, CD3, FOXP3, and PD1. These results demonstrated valid substitution of chromogenic staining for single-plex IF and that sequential multiplexing on a single tissue section can be representative of the single-plex staining for several antibodies, but only if the effect of order in the multiplex protocol is considered. The order should be tested for each antibody combination used, to enable high concordance between single-plex and multiplex data.
In conclusion, an optimised multiplex IF protocol was devised, in which correlation between multiplex antibody staining and single-plex staining was maximised. Knowledge that multiplex protocols can reach concordance with clinically validated singleplex chromogenic assays is of great importance. Clinical pathology presently relies on single-plex chromogenic staining, whilst multiplex IF will be important as personalised medicine increasingly entails assessment of multiple biomarkers, preferably simultaneously. Multiplex IF will conserve limited clinical material and enable spatial resolution of co-localised antigens to facilitate numeration of complex immune cell subpopulations, which require several markers for their identification. These data will increase our understanding of the molecular mechanisms involved in the anti-tumour immune response.
HKA, GNJ, AL, AS are employees and shareholders of AstraZeneca. Collaborative Research Agreement with Perkin- Elmer and Definiens (of relevance to this manuscript). WH is an employee and shareholder of Abcam PLC and former employee of AstraZeneca. JS, JA, JJ, CS, and RB have no conflicts.
Citation: Syed J, Ashton J, Joseph J, Jones GN, Slater C, Sharpe A, et al. (2019) Multiplex Immunohistochemistry: The Importance of Staining Order When Producing a Validated Protocol. Immunotherapy (Los Angel) 5: 157. Doi: 10.35248/2471-9552.19.5.157
Received: 04-Dec-2019 Accepted: 18-Dec-2019 Published: 25-Dec-2019
Copyright: © 2019 Syed J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.