Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2024)Volume 17, Issue 3
Spinacia oleracea is a very famous medicinal plant that bears a lot of biological activities. The high pressure in the pulmonary arteries, which is a dangerous unhealthy condition leading to heart failure and finally death, is called Pulmonary Arterial Hypertension (PAH). Hence, in the current study, there was an attempt to probe into possible active constituents of spinach with the use of network pharmacology and molecular docking in the treatment of PAH. The results identified the common targets between spinach constituents and PAH-related proteins, including hub targets such as Epidermal Growth Factor Receptor (EGFR), Peroxisome Proliferator-Activated Receptor Gamma (PPARγ), Sarcoma Proto-Oncogene (SRC) and Matrix Metalloproteinase 9 (MMP9). Pathway analysis revealed association with lipid metabolism, atherosclerosis and the Interleukin 17 (IL-17) signaling pathway. Molecular docking showed strong binding affinity between patuletin, spinacetin and the PAH-associated targets. These results have generated insight into the molecular mechanism underlying possible PAH therapies by spinach constituents.
Spinach; Spinacia oleracea; Pulmonary arterial hypertension; Network pharmacology; Molecular docking
The common names for Spinacia oleracea are "spinach" and "paalak". It is a dark green color leafy vegetable, originally annual and rarely biennial plant belonging to the Chenopodiaceae family (Table 1) [1]. There is broad categorization of spinach corresponding to their leaf texture. Alternate leaf structure with the lower having a one very long petiole, variously lobed with lobes in acute triangle shape. The texture of leave is smooth on both the sides. Straight up erected to a length height of 30-60 cm high, smooth texture and round diameter, piped, succulent stems, at some parts, the tips are red in colour. Other potential health benefits of spinach consumption include improved control in blood glucose, reduced risk of cancer and improved bone health [2]. Nutritionally, spinach is made up mostly of water 91.4%, protein 2.9%, carbohydrate 3.6% and fat 0.4%. The lipid fraction is primarily composed of mono and polyunsaturated fatty acids while saturated fatty acids are present only in minute amounts. The 100 g serving of spinach contains higher amounts of magnesium, potassium and iron and covers 20%, 16% and 15%, respectively, of their recommended dietary allowance [3]. Spinach is incredibly rich in flavonoids. So many flavonoids, such as quercetin, myricetin, spinacetin, luteolin, jaceidin, patuletin, lutein epoxide, neoxanthin, glucuronic acid 3,5,7,30,4'pentahydroxi-6-methoxiflavone, pheophytin b, neoluteinare, have been reported to be found in spinach [4]. Spinach is rich in the content of phenolic compounds and carotenoids. The polyphenols detected from the spinach and spinach derived extracts are para-coumaric acid, ortho-coumaric and ferulic acid. Spinach also demonstrates the presence of different carotenoids like lutein, 90-(Z)-neoxanhin, violaxanthin and β-carotene [5]. Spinach contains a high level of vitamin A, C, E and K. In addition, it is equally rich in folic acid. With these health-promising compounds, spinach and its extracts are rich in various minerals such as calcium, manganese magnesium, iron, copper, zinc, potassium and phosphorus [6]. All ailments and disorders are usually associated with administering drugs that are in use today. Most of these drugs seem to have adverse, irreversible side effects, which include nausea, aversion, constipation, dizziness and even weight gain. Because of this, functional meals have come to gain popular use as substitutes for the cure and prevention of lifestyle disorders [7].
| Kingdom | Plantae |
|---|---|
| Division | Magnoliophyta |
| Class | Magnoliopsida |
| Order | Caryophyllales |
| Family | Chenopodiaceae |
| Genus | Spinacia |
| Species | oleracea |
Table 1: Scientific classification of Spinacia oleracea.
One of the five varieties of Pulmonary Hypertension (PH), PAH is characterized by increased pulmonary arterial pressure and vascular resistance and severe, progressive symptoms [8]. A number of characteristic features define PAH, which is a complex and multidimensional disease comprising inflammation, decreased angiogenesis, metabolic changes and extravascular deposition of collagen [9,10]. Sustained elevation of pulmonary-arterial pressure of greater than 25 mm Mercury (Hg) at rest or greater than 30 mm Hg with exercise, with a mean pulmonary-capillary wedge pressure and left ventricular end-diastolic pressure of less than 15 mm Hg [11]. The histologic appearance of lung tissue in each of these conditions is similar; intimal fibrosis, increased medial thickness, pulmonary arteriolar occlusion and plexiform lesions predominate [12].
One important strategy for the treatment of diseases is to develop drugs that can act on multiple proteins involved in the symptoms of a disease. Building multiple networks elucidating gene targets, diseases and drugs and network pharmacology researches applying systems biology and computational methods on Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways would be helpful in the generation of such drugs [13,14]. In addition, this molecular docking methodology computes the energies of binding between ligands and receptors, which helps in choosing the appropriate binding modes [15].
In this respect, the present work identifies possible targets and mechanisms of action of Spinacia oleracea active components against PAH using network pharmacology and molecular docking. Such an approach would reveal potential therapeutic targets against PAH and provide a molecular basis for the potential application of spinach in the treatment of this disease.
Screening of phytoconstituents of Spinacia oleracea
The chemical constituents in Spinacia oleracea was acquired from the database of Indian Medicinal Plants, Phytochemistry and Therapeutics (IMPPAT) (https://cb.imsc.res.in/imppat/); 32 compounds were collected. Using canonical Simplified Molecular Input Line Entry System (SMILES), the bioactive compounds were obtained using Drug-likeness and Lipinski violation 0^SwissADME (http://www.swissadme.ch/) databases respectively [16,17]. We used the ProTox webserver (https://tox.charite.de/protox3/), to predict the toxicity of these compounds, which ultimately led to the selection of 9 active constituents [18].
Obtaining Spinacia oleracea related target genes
The SMILES file and canonical molecular structure were acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Subsequently, target genes were obtained with the help of the SwissTargetPrediction (http://www.swisstargetprediction.ch/) and Search Tool for Interactions of Chemicals (STITCH) (http://stitch.embl.de/) databases [19,20]. Target prediction in each of these databases was limited to Homo sapiens and thereafter, the targets selected in all of them were pooled with duplicate values removed for analysis into target genes.
Obtaining pulmonary arterial hypertension related target genes
The GeneCards database was searched using the keywords "Pulmonary arterial hypertension," and target genes linked with PAH were found (https://www.genecards.org/) [21]. The maximum score value obtained is 337.15; the minimum score is 0.32. The target with a score>5 is set as the potential target of PAH.
Obtaining potential common targets
A Venn diagram was made to import the prospective targets into the web tool Venny 2.1 and get the common potential targets of PAH and the seven active compounds (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
Protein-Protein Interactions (PPIs) and network analysis
To determine the plant target gene interaction networks for PAH, Homo sapiens was set as the single organism and an interaction network was then constructed using the search tool of version 12.0 of the retrieval of interacting genes and proteins tool, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/). The PPI network was constructed with a minimum required interaction score of>0.400 [22]. After the files were loaded into Cytoscape v.3.10.2, the hub genes were found using Cytoscape's cytoHubba plugin on the degree method and higher-degree nodes [23-25].
Pathways and Gene Ontology (GO) enrichment analysis
Gene Ontology (GO) analysis is currently a popular way to analyse genomic data, particularly large-scale transcriptome data. Potential targets were analysed for GO functional enrichment in 3 groups; Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) [26,27]. The Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/tools.jsp) and ShinyGO software version 0.76 (http://bioinformatics.sdstate.edu/go76/) was used to explore pathways and diseases related to the hub genes in the KEGG (https://www.genome.jp/kegg/) respectively [28,29]. Results of gene set enrichment with a p-value of less than 0.05 were considered statistically significant.
Molecular docking
The structure of seven active constituents was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) i.e, spinacetin, patuletin, dihydro-beta-ionone, beta-ionone, 5,3',4'-trihydroxy-3-methoxy-6,7-methylenedioxyflavone, quercetin and thioctic acid. The Schrödinger Maestro v13.5 software was used to create the Three-Dimensional (3D) structures of all ligands. The ligands were produced at physiological pH (7.0 ± 2.0) using the LigPrep and Epik modules from the Schrodinger's suite. After the ligands were transformed into their 3D structures, ionization was performed using the Optimized Potentials for Liquid Simulations 4 (OPLS4) force field to generate tautomeric states [30,31]. Four proteins that met the criteria were Epidermal Growth Factor Receptor (EFGR), Peroxisome Proliferator Activated Receptor Gamma (PPARG), Proto Oncogene or Non-Receptor Tyrosine Kinase (SRC) and Matrix Metallopeptidase 9 (MMP9). The 3D crystal structures of target proteins were selected and acquired from the Protein Data Bank (PDB) (https://www.rcsb.org/). For the EFGR target, protein structure with PDB ID: 3POZ; PDB ID: 8BF1 for the PPARG target; PDB ID: 1FMK for the SRC target and PDB ID: 1ITV for the MMP9 target. The protein preparation method was employed for removal of water molecules from the crystal structures, addition of missing hydrogen atoms and closure of loop gaps and modification of the side chain protonation states. Charges, bond orders and hydrogens were assigned to the heavier atoms. Water was removed and selenomethionines were converted into methionines. The prepared protein was then fragmented before being processed further for grid generation. The co-crystallized enzyme ligand was then covered by a receptor grid to expose the binding site [32].
In Glide of Schrödinger-Maestro v 13.5, following the High-Throughput Virtual Screening (HTVS) as a validation step, non-cis/trans amide bonds were penalized during XP flexible ligand docking [33]. The partial charge cutoff and van der Waals scaling factor for ligand atoms were selected to be 0.15 and 0.80, respectively [34]. For each of the functional groups identified, there is bias sampling of torsions and the docking score is enhanced with the application of Epik state penalties [35]. Final scoring was according to energy-minimized poses and represented as a docking score. For each ligand, it was identified with the best-docked site for which the value of the docking score was lowest.
Identification and filtration of active constituents of Spinacia oleracea
The ligands were searched, identified, screened and duplicates removed to identify seven active compounds with a DL ≥ 0.18 and violation of the Lipinski rule=0 (Table 2).
| Compound | Molecular weight | Lipinski violations | Drug likeness | 2D structure |
|---|---|---|---|---|
| Spinacetin | 346.29 | 0 | 0.45 | 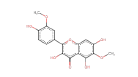 |
| Patuletin | 332.26 | 0 | 0.5 | 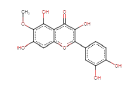 |
| Dihydro-beta-ionone | 194.31 | 0 | 0.19 | 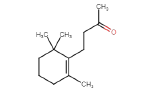 |
| beta-Ionone | 192.3 | 0 | 0.29 | 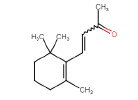 |
| 5,3',4'-Trihydroxy-3-methoxy-6,7-methylenedioxyflavone | 344.27 | 0 | 0.36 | 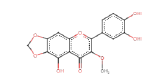 |
| Quercetin | 302.24 | 0 | 0.52 | 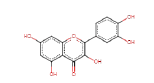 |
| Thioctic acid | 206.33 | 0 | 0.35 | 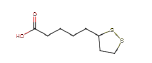 |
Table 2: Active constituents of Spinacia oleracea and their properties.
Screening of potential PAH for Spinacia oleracea
The potential targets of PAH and Spinacia oleracea were obtained from various databases. Possible targets associated with PAH were obtained 2845 in number after removing and merging the duplicate values from the GeneCards database. Target genes for Spinacia oleracea were obtained in the count of 328 from STITCH and SwissTargetPrediction databases. Common targets of PAH and Spinacia oleracea were obtained 172 in number through intersection via Venn diagram (Figure 1).
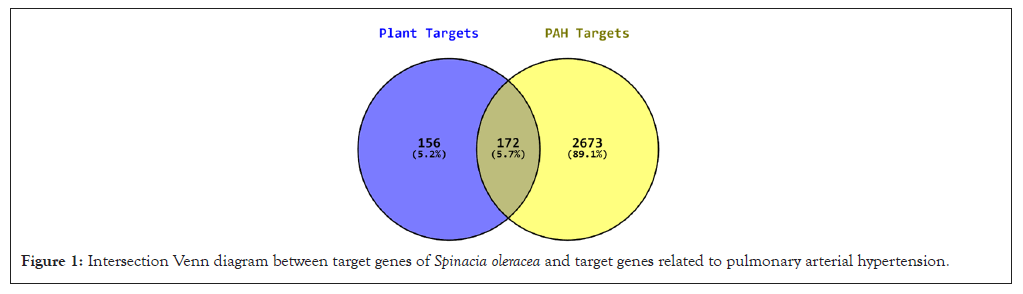
Figure 1: Intersection Venn diagram between target genes of Spinacia oleracea and target genes related to pulmonary arterial hypertension.
PPI network and hub genes analysis
The STRING version 12.0 database was utilized to import the 172 common targets in order to build the PPI network and display the correlation among the possible targets. The PPI network has 171 nodes and 268 edges, with an average node degree of 3.13 and a clustering coefficient of 0.485, according to the results. PPI enrichment had a p-value of less than 1.0×10-16. After that, a network diagram of the hub targets of Spinacia oleracea and PAH was created. This was done by analysing the PPI network using the degree method and the cytoHubba add-on plugin of the Cytoscape software. The results showed that the top 10 hub targets were IL-6, AKT Serine/Threonine Kinase 1 (AKT1), Prostagladin-Endoperoxide Synthase 2 (PTGS2), Epidermal Growth Factor Receptor (EGFR), Estrogen Receptor 1 (ESR1), Peroxisome Proliferator Activated Receptor Gamma (PPARG), Proto Oncogene or Non-receptor Tyrosine Kinase (SRC), Matrix Metallopeptidase 9 (MMP9), Heat Shock Protein 90 Alpha Family Class A Member 1 (HSP90AA1) and C-X-C Motif Chemokine Ligand 8 (CXCL8) (Figures 2-4).
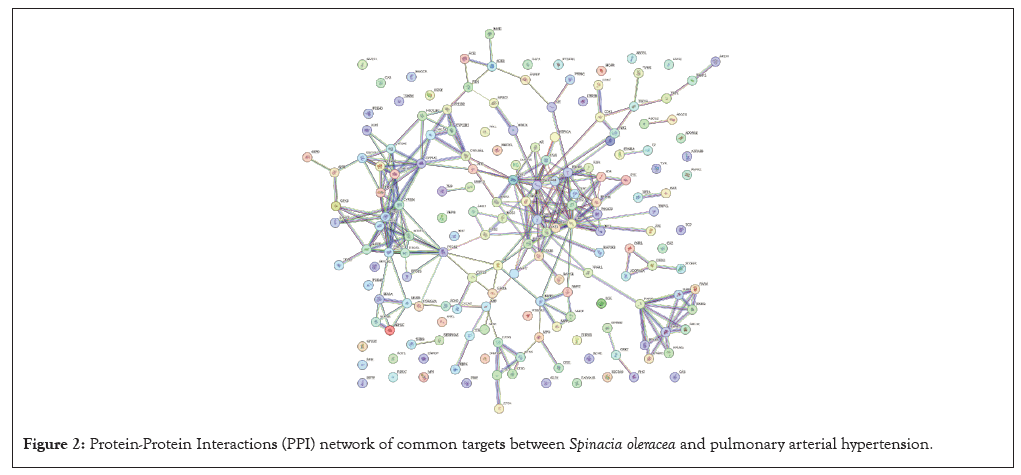
Figure 2: Protein-Protein Interactions (PPI) network of common targets between Spinacia oleracea and pulmonary arterial hypertension.
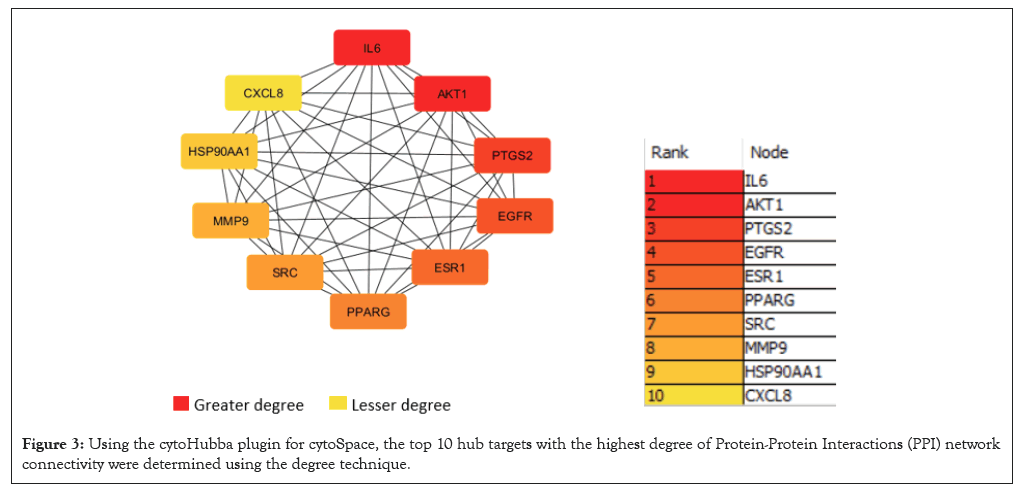
Figure 3: Using the cytoHubba plugin for cytoSpace, the top 10 hub targets with the highest degree of Protein-Protein Interactions (PPI) network connectivity were determined using the degree technique.

Figure 4: Bar chart of top 10 hub genes by degree method with scores.
Gene ontology and KEGG pathway enrichment analysis
KEGG pathway and gene ontology enrichment analyses were performed using the online tool DAVID to elucidate and demonstrate the molecular mechanism. This GO enrichment study has three main divisions: Molecular Functions (MF), Cellular Components (CC) and Biological Processes (BP). As per the BP, the large majority of the target genes are predominantly enriched for response to hormone, response to lipids, response to organic cyclic compounds, response to organonitrogen compounds, cellular response to oxygen-containing compound, response to oxygen compound, response to endogenous stimulus, regulation of the programmed cell death, cell death, regulation of localization, etc. The CC showed that most of the target genes were significantly enriched in membrane rafts, membrane microdomains, receptor complexes, the cell surface, the perinuclear region of the cytoplasm, the organelle envelope, the side of the membrane, extracellular reticulum, vesicle, caveola and others. Results obtained showed that these central targets were associated with functions such as nuclear receptor activity, ligand-activated transcription factor activity, steroid hydroxylase activity, Heme binding, monooxygenase activity, zinc ion binding, protein kinase activity, oxidoreductase activity, anion binding, cation binding, metal ion binding and so on. Moreover, the results of KEGG pathway enrichment analysis indicated that IL-17 signaling pathway, fluid shear stress and atherosclerosis, lipid and atherosclerosis, relaxin signaling pathway, metabolic pathways and so on correlated with the predicted hub targets for PAH (Figure 5).
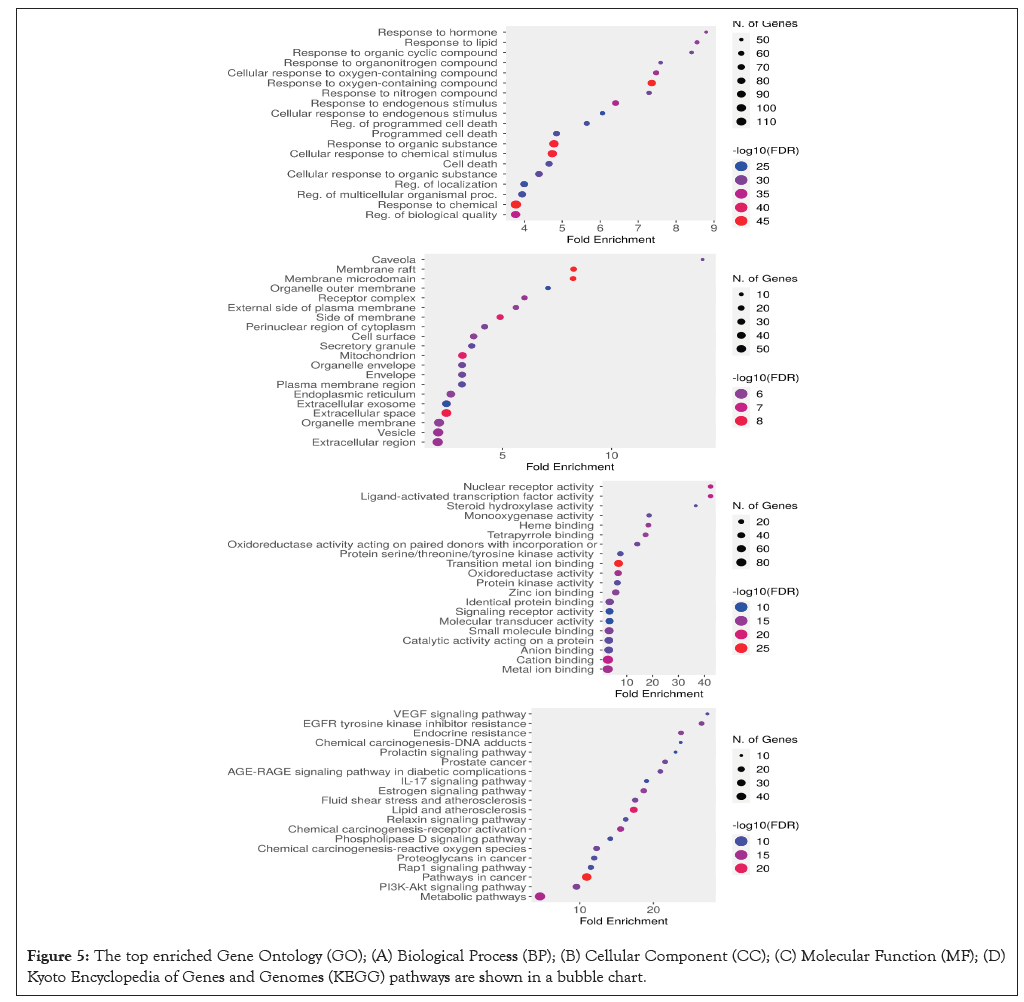
Figure 5: The top enriched Gene Ontology (GO); (A) Biological Process (BP); (B) Cellular Component (CC); (C) Molecular Function (MF); (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are shown in a bubble chart.
The hierarchical clustering tree condenses association information for the important pathways presented on the enrichment chart. Larger dots indicate more significant p-values and the pathways with a large number of common genes are located together (Figure 6).
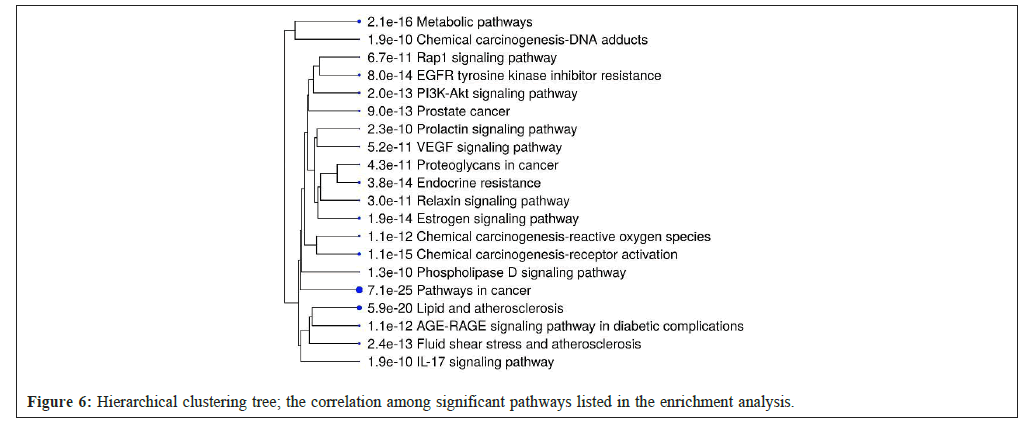
Figure 6: Hierarchical clustering tree; the correlation among significant pathways listed in the enrichment analysis.
Molecular docking analysis
Screening of the four hub targets for PAH against 7 active constituents whose binding energy was successfully predicted using docking analysis. All of the top active constituents demonstrated good binding and a high degree of matching with the target protein. Among all of them, patulein and spinacetin showed a good docking score against the four hub targets (Table 3).
| SL.No | Hub targets | PDB ID | Constituents | Docking score (Kcal/mol) | Interacting residues |
|---|---|---|---|---|---|
| 1 | EFGR | 3POZ | Patuletin | -7.498 | GLU:872, TYR:891, ARG:889, ILE:886, SER:885. |
| Spinacetin | -6.561 | ARG:889, SER:885, GLU:872. | |||
| 2 | PPARG | 8BF1 | Patuletin | -9.265 | GLU:259. |
| Spinacetin | -6.561 | SER:342, PHE:264, ASP:260,GLU:259,GLN:273. | |||
| 3 | SRC | 1FMK | Patuletin | -8.342 | SER:248, GLU:320, GLN:144, GLU:146. |
| Spinacetin | -6.707 | GLN:144, PHE:150. | |||
| 4 | MMP9 | 1ITV | Patuletin | -8.043 | PRO A:143, MET B:141. |
| Spinacetin | -7.321 | PRO A:143, MET B:141. |
Table 3: Docking score of patuletin and spinacetin with hub targets.
Results showed that the EFGR Hub target, PDB ID: 3POZ, had a score of with patulein.
-7.498 Kcal/mol, having hydrogen bonds in the following amino acids: GLU872, ARG889, TYR891, ILE886 and with pi-cation bridge at ARG889 amino acid. The spinacetin with EFGR Hub target with PDB ID: 3POZ had a score of -6.561Kcal/mol, with hydrogen bonds in the following amino acids: GLU872, ARG889, SER885 and Pi-cation bridge at the ARG889 amino acid (Figure 7A and 7B).
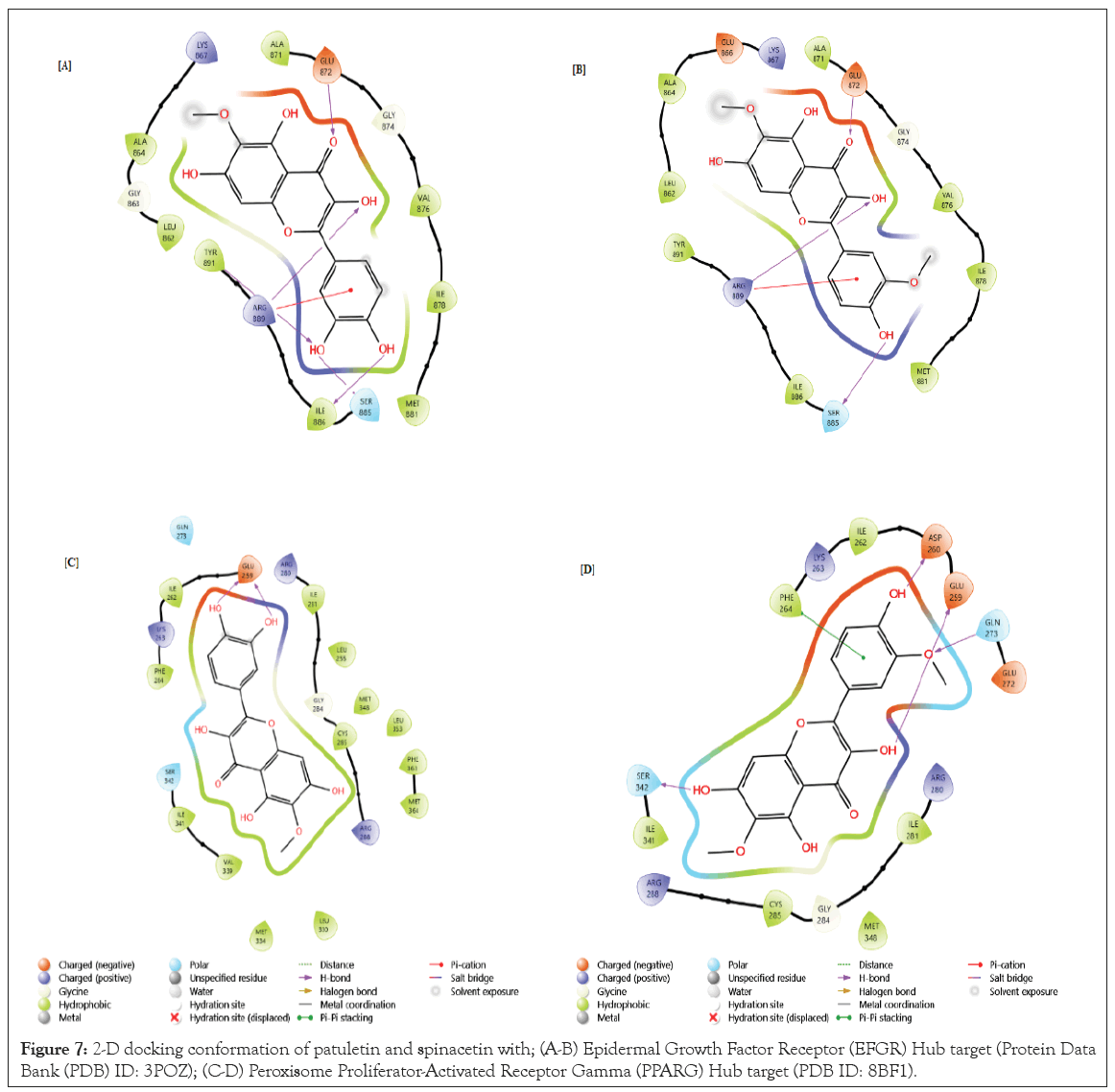
Figure 7: 2-D docking conformation of patuletin and spinacetin with; (A-B) Epidermal Growth Factor Receptor (EFGR) Hub target (Protein Data Bank (PDB) ID: 3POZ); (C-D) Peroxisome Proliferator-Activated Receptor Gamma (PPARG) Hub target (PDB ID: 8BF1).
The results showed that patulein with PPARG Hub target (PDB ID: 8BF1) possessed a score of -9.265Kcal/mol, with hydrogen bonds present in the subsequent amino acids: GLU259, GLU259. The spinacetin with PPARG Hub target (PDB ID: 8BF1) possessed a score of -6.561Kcal/mol, with hydrogen bonds present in the subsequent amino acids: ASP260, GLN259, GLN273, SER342 and pi-pi stacking bridge at PHE264 amino acid (Figure 7C and 7D).
The results showed that SRC Hub target (PDB ID: 1FMK) with patulein possessed a score of
-8.342 Kcal/mol, with hydrogen bonds present in the subsequent amino acids: SR248, GLN144, GLU146, GLU320 and the spinacetin with SRC Hub target (PDB ID: 1FMK) possessed a score of -6.707Kcal/mol, with hydrogen bonds present in the subsequent amino acids: PHE150, GLN144 (Figure 8A and 8B).
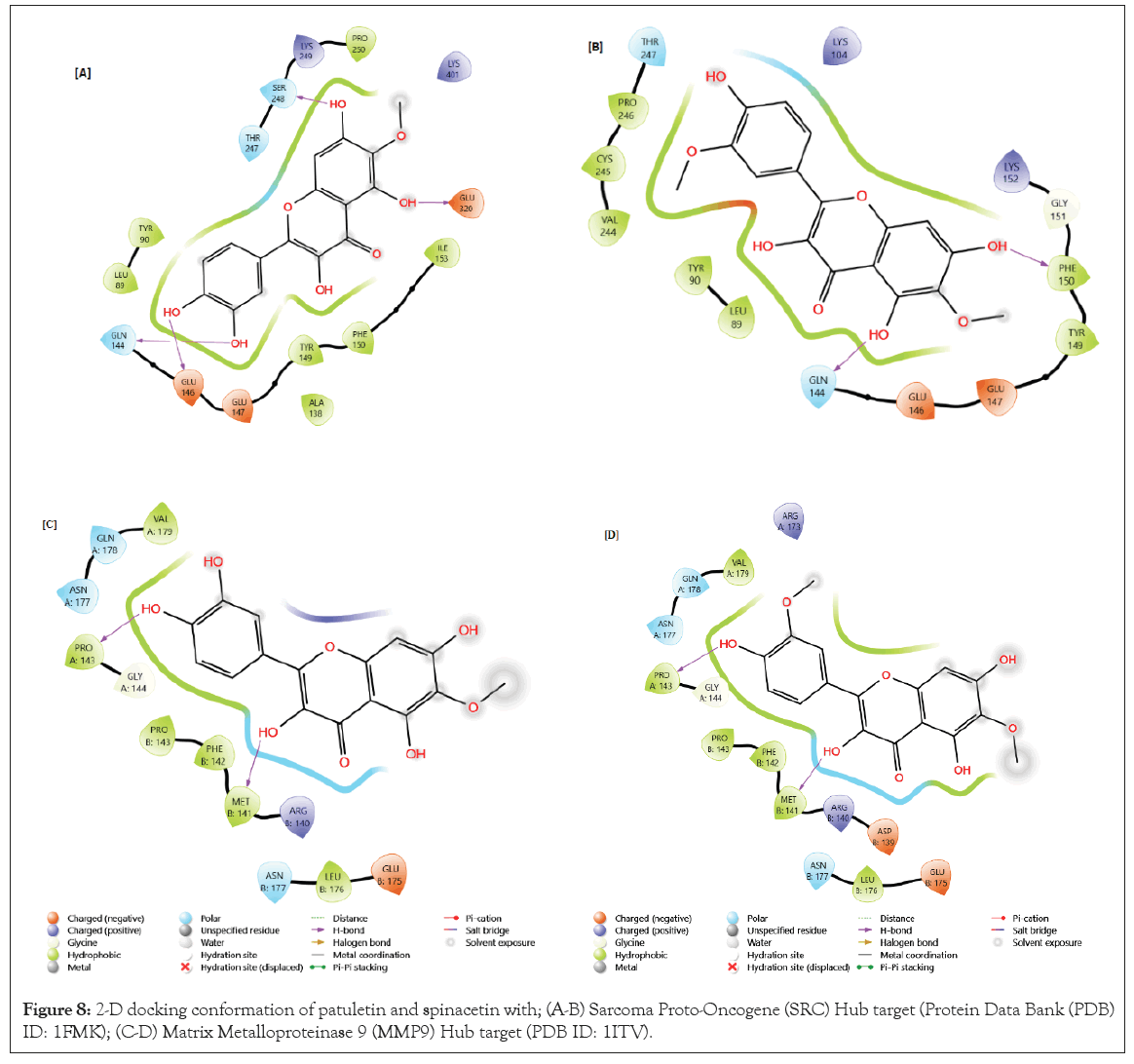
Figure 8: 2-D docking conformation of patuletin and spinacetin with; (A-B) Sarcoma Proto-Oncogene (SRC) Hub target (Protein Data Bank (PDB) ID: 1FMK); (C-D) Matrix Metalloproteinase 9 (MMP9) Hub target (PDB ID: 1ITV).
The results showed that patulein with MMP9 Hub target (PDB ID: 1ITV) possessed a score of
-8.043Kcal/mol, with hydrogen bonds present in the subsequent amino acids: PRO A143, MET B141. The spinacetin with MMP9 Hub target (PDB ID: 1ITV) possessed a score of -7.321 Kcal/mol, with hydrogen bonds present in the subsequent amino acids: MET B141, PRO A143 (Figure 8C and 8D).
This work was aimed at a systematic exploration of the possible targeting of active ingredients in the treatment of PAH using network pharmacology and molecular docking methodology. We had unique targets between plant targets and PAH targets from our database search. In addition, PPI analysis revealed 10 hub targets among the 172 common targets: IL6, AKT1, PTGS2, EGFR, ESR1, PPARG, SRC, MMP9, HSP90AA1 and CXCL8. The results show that these ten hub targets are mainly associated with the pathways of metabolic pathways, EGFR tyrosine kinase inhibitor resistance, Phosphoinositide 3-Kinase-Protein Kinase B (PI3K-Akt) signalling pathway, Vascular Endothelial Growth Factor (VEGF) signalling pathway, endocrine resistance, phospholipase D signalling pathway, lipid and atherosclerosis, IL-17 signalling pathway.
Interestingly, the lipid pathways and atherosclerosis biological process were indicated to be the primary targets of plant active ingredients, despite no direct relatedness to lipid metabolism or accumulating fat in the artery. Putatively, lipid pathways and atherosclerosis may seem related targets due to cross-talk of biological processes and signaling pathway interconnections involved in the context of PAH. Consequently, PAH is a complicated pathophysiological condition with many alterations in the homeostasis of the pulmonary vascular system that invoke a variety of cellular and molecular responses. While the top plant targets identified are not seemingly directly related to lipid metabolism, they could be taking part in molecular interactions or signaling cascades that are indirectly related to this pathway. It was carried out to predict and validate, on a molecular basis, the docking of plant compounds into the binding sites of 10 potential hub targets for PAH, including IL6, AKT1, PTGS2, EGFR, ESR1, PPARG, SRC, MMP9, HSP90AA1 and CXCL8. Encouragingly, the results showed patulein and spinacetin binding onto inside of these central targets well. It is worth noting that the lower the binding energy between the ligand or drug and a protein, the higher the affinity estimated.
EGFR is a hub target that plays a central role in the pathogenesis of PAH. Activation of the transmembrane receptor tyrosine kinase, EGFR, occurs after binding to its ligands, such as epidermal growth factor and transforming growth factor-α. In PAH, abnormal vascular remodelling in the pulmonary arteries mediates a generic response that leads to increased pulmonary vascular resistance. Once activated, EGFRs can contribute to smooth muscle cell proliferation, endothelial dysfunction and fibrosis— some of the hallmarks associated with vascular remodeling.
PPARG is a ligand-activated transcription factor that controls genes modulating pathways, which contribute to vascular tone and remodelling. It has anti-inflammatory effects, inhibiting factors implicated in PH, such as vascular cell adhesion molecule, IL-6 and monocyte chemoattractant protein. Activation of PPARG reduces endothelin 1 levels and confers resistance to apoptosis on endothelial cells.
SRC target plays a vital role in the potassium channel function of human pulmonary artery smooth muscle cells. A response to changes in oxygen tension. Target, therefore, is a potential point of therapeutic intervention. Clinical use of a SRC inhibitor has led to partially reversible pulmonary hypertension. Familial pulmonary hypertension involves the transforming growth factor-beta signalling pathway, endoglin receptor causing abnormal vascular remodeling.
MMP-9, has been shown to be a zinc-dependent endopeptidase that plays a central role in vascular remodeling in pulmonary arteries. Higher levels of MMP-9 have been linked with cardiac and vascular changes. Induction of the proteolytic pathways, including MMP-9, results in an over-proteolysis of extracellular proteins considered significant for arterial remodelling. Although by itself, MMP-9 cannot be used as a diagnostic marker, plasma levels are directly correlated with the severity of disease.
Therefore, this network pharmacology-based study is helpful in explaining the mechanism of action for active ingredients, probable target genes and link pathways regarding the treatment of PAH, thus laying a foundation for further experimental verification. Of course, some information here is really interesting; however, further study and research projects should be conducted to fully understand the potential of Spinacia oleracea in order to prove its medicinal value.
The current study integrates network pharmacology with molecular docking analysis to illustrate Spinacia oleracea's function in the treatment process of PAH. Network pharmacology and molecular docking approach were employed in the current study for probing the mechanism underlying the identified phytochemical constituents and their interaction with target proteins with PAH. It also showed several signalling pathways linked with the therapeutic mechanism of Spinacia oleracea in the KEGG pathways analysis. These pathways include those related to PAH, metabolic pathways, EGFR tyrosine kinase inhibitor resistance, PI3K-Akt signalling pathway, VEGF signalling pathway, endocrine resistance, phospholipase D signalling pathway, lipid and atherosclerosis and IL-17 signalling pathway. The binding affinities, including strong interactions to a panel of hub targets and key residue-specific interactions demonstrate how these constituents may be used as building blocks for further development and improvement. Additional studies both in vivo and in vitro are needed to elaborate the roles of spinacetin and patuletin in the development of PAH, to confirm and progress these findings. This work therefore lays the basis for future studies that may go on to result in drug discovery and development of more potent and specific therapies. This work thus lays the groundwork for future investigations into drug discovery and development, which will lead to more potent and specific therapies.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Reddy YV, Varma TT, Anand V (2024). Network Pharmacology and Molecular Docking Reveals Spinacia oleracea Constituents as Potential Pulmonary Arterial Hypertension (PAH) Therapeutics. J Proteomics Bioinform. 17:672.
Received: 16-Aug-2024, Manuscript No. JPB-24-33501; Editor assigned: 19-Aug-2024, Pre QC No. JPB-24-33501 (PQ); Reviewed: 02-Sep-2024, QC No. JPB-24-33501; Revised: 09-Sep-2024, Manuscript No. JPB-24-33501 (R); Published: 16-Sep-2024 , DOI: 10.35248/0974-276X.24.17.672
Copyright: © 2024 Reddy YV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.