
Immunotherapy: Open Access
Open Access
ISSN: 2471-9552

ISSN: 2471-9552
Research Article - (2019)Volume 5, Issue 1
Recent research has revealed that exosomes affect several aspects of organism physiology and development, as well as disease processes, through intracellular communication. However, the biological roles of exosomes in exosomesecreting cells have remained largely unexplored. In this study, we demonstrate that Schwann cell (SC)-derived exosomes (EXOSC) promote cell survival among motor neurons (MNs), with MN viability exceeding 80% at days in vitro (DIV) 14. Prevention of MN cell death by EXOSC was achieved by blocking the caspase-3 cell death pathway, which was confirmed by comparing the number of activated-caspase3+ cells according to the presence versus absence of EXOSC. The attenuation of exosome secretion from SCs by treatment with GW4869 resulted in increased MN cell death, regardless of SC viability. Together, these findings enhance our understanding of exosome biology and provide valuable new insights into exosomes as a potential therapeutic agent in nervous system disorders.
Cell survival; Exosome therapy; Motor neurons; Nerve regeneration; Peripheral nervous system; Schwann cell-derived exosomes
In adult mammals, peripheral nervous system (PNS) neurons regenerate relatively efficiently, whereas central nervous system (CNS) neurons cannot regenerate. The difference in regenerative capacity between CNS and PNS neurons has been attributed to extrinsic signals, and to intrinsic growth ability and the inhibitory glial environment. PNS neurons possess the fundamental and conserved ability to regenerate their axons following injury, in a process mainly controlled by the PNS glial cells surrounding neurons and Schwann cells (SCs). Several studies have reported the regenerative ability of SCs [1-6]. After axotomy, SCs undergo a rapid reprogramming process that includes macrophage activation and morphological transformation from de-differentiating into non-myelinating repair cells that provide the signals and cues necessary for recovery of injured neurons, axon regeneration, and reinnervation [1,2]. Although these complex intrinsic and extrinsic processes can restore damaged neurons, the SC repair system is efficacious only for injuries less than 3 cm in size [7,8]. To achieve functional recovery, patient age and delays in repair after axotomy must be fully considered. Recent studies have confirmed that SCs are a remarkable cell regeneration resource; however, little is known regarding the molecular mechanism underlying spontaneous axon regeneration. More advanced strategies are required before SCs can be applied for complete restoration of damaged neurons.
Intracellular communication, a hallmark of living organisms, is mediated through cell-to-cell interactions or the secretion of molecules such as growth factors, cytokines, adhesin molecules, chemokines, and/or shed molecules. Secretome can be released as either soluble molecules or within extracellular vesicles (EVs). Exosomes, a type of cell-derived EV, have recently emerged as critical mediators and promoters of intracellular communication between donor and recipient cells, through delivery of certain substances in healthy and diseased nervous systems [9-12]. Nanosized exosomes carrying nucleic acids, proteins, lipids, and RNAs originate from the endocytic compartment of the donor cell and deliver genetic cargo to the cytoplasm of the target cell, thereby providing a unique mechanism for intracellular transport. Neuron-derived exosomes can influence synaptic plasticity, synaptic strength, and the viability of neighboring cells [13-15], and CNS glial cell-derived exosomes are involved in the regulation of myelination, alleviation of oxidative stress, neuroprotection, and neurotransmission, suggesting that nervous system exosomes could have significant clinical applications as biomarkers or therapeutic agents. Little is currently known regarding the membrane vesicles secreted by glial cells in the PNS. SC-derived exosomes promote axon regrowth in injured dorsal root ganglia (DRG) [16] and also play a role in the inhibition of CNS neuron inflammation [11]. In the future, exosomes may play an essential role as a neurorestorative resource in the treatment of neurodegenerative disease.
Recently, motor neuron (MN) cell viability was found to be enhanced by SC-conditioned medium [17,18]. However, the precise molecular mechanism by which factors secreted from SCs confer neuroprotection, and whether these factors originate from soluble proteins or vesicles, remains unclear. In this study, we treated MNs with SC-derived exosomes (EXOSC) to explore their role in preserving MN cell viability.
Cell preparation
All procedures pertaining to the acquisition of biological samples were approved by the Institutional Animal Care and Use Committee (IACUC) of Hanyang University (HY), and all experiments were conducted in accordance with the guidelines and regulations stipulated by the committee. All mice were purchased from DBL (Eumseong, Korea) in agreement with the research protocols approved by the IACUC of HY.
Primary SC culture
SC culturing was performed as previously described [18,19]. Briefly, sciatic nerves from postnatal day 4 mice were harvested in Ca2+- and Mg2+-free phosphate-buffered saline (PBS, Gibco) and incubated with a mixture of 2.5% trypsin (Gibco) and 1 mg/mL collagenase A (Roche) in a 37°C water bath for 30 min. The sciatic nerve solution was washed with high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Welgene) containing 10% fetal bovine serum (FBS, Gibco) and then suspended in SC culture medium consisting of DMEM with 10% FBS, 4 mM L-glutamine (L-gln, Invitrogen), 0.5 μM forskolin (Sigma), 2 ng/mL human heregulin beta-1 (Sigma), and 1% penicillin/streptomycin (P/S, Gibco). SC suspension was seeded on coverslips pre-coated with 10 μg/mL poly-Dlysine (PDL, Sigma). After 3 days, complement-mediated cytolysis was performed to remove fibroblasts from the SC culture, as previously described [18,20]. Briefly, cells were treated with DMEM containing 20 mM hydroxyethyl-piperazineethanesulfonic acid (HEPES) buffer (T and I), 10% FBS, 4 mM L-gln, and 1% P/S) for 5 min before washing with Ca2+- and Mg2+- free Hank’s balanced salt solution (HBSS, Invitrogen) containing 20 mM HEPES. Cells were then treated with 4 ng/mL anti-mouse CD90 (Bio-Rad) in DMEM containing 20 mM HEPES buffer (T and I), 10% FBS, 4 mM L-gln, and 1% P/S) at 37°C for 2 h. Complement mediated cytolysis was terminated by washing cells with 20 mM HEPES. At 48 h prior to the collection of SC-conditioned medium, SC culture medium was completely replaced with fresh culture medium containing exosome-depleted FBS (EXO-FBS; System Bioscience).
Primary MN cell culture
MN culturing was performed as described previously [18,20]. 12 fetuses) were harvested in Ca2+- and Mg2+-free HBSS containing 1% Toll-like receptor (TLR) trypsin (Worthington) for 15 min at 37°C, followed by treatment with 1% trypsin inhibitor (Sigma). Dissociated spinal cord pieces were then incubated in an immunopanning dish pre-coated with p75NTR antibody (Abcam) at room temperature for 45 min to collect purified MNs. To remove p75NTR-negative cells and nerve fragments, the immunopanning dish was washed with neurobasal medium (NB; Gibco) containing GlutaMAX × 1 (Gibco). MNs bound to the bottom of the immunopanning dish were detached with depolarization solution and gently harvested in SC–MN co-culture medium [NB containing 1% FBS, 1% P/S, 1× B27 supplement (Gibco), 0.5 μM forskolin, 1 mg/mL bovine pituitary extract (Gibco), and 10 μg/mL brain-derived neurotrophic factor (BDNF, Gibco). EXOSC were added to cell culture medium and their concentration was maintained throughout culturing until analysis. Z-DEVD-FMK004 (R and D Systems) or dimethyl sulfoxide (DMSO) vehicle (Sigma) was added to culture media at 3 days in vitro (DIV) and the protein concentration was maintained throughout culturing until fixation for analysis.
Transwell culture
To assess the viability of MNs growing in a Transwell (Corning), purified MNs seeded on coverslips pre-coated with 0.5 mg/mL poly-ornithine hydrobromide and 2.5 mg/mL laminin were placed in the bottom compartment of the Transwell. Transwell inserts were precoated with 10 μg/mL PDL solution for 2 ~ 3 h at 37°C and washed three times with PBS before seeding with SCs. At 3 days after MN plating in the bottom compartment of the Transwell, MNs were treated with or without EXOSC or SCs in the upper Transwell insert.
For SC–MN Transwell co-culture, both cell types were cultured on the same day, and combined after 3 days.
Exosome purification
Exosome purification was performed as described previously [21], with minor modifications. Briefly, SC-conditioned medium was collected at DIV 5 and centrifuged to remove cell debris at 3,000 rpm for 15 min. The supernatant including exosomes was carefully harvested and kept on ice prior to performing exosome separation using the ‘H’method. To isolate exosomes, the flow ratio of sample:buffer:magnification was set at 5:95:75. Exosome-sized particles were automatically separated from the other particles; all samples were maintained at 4°C during exosome purification. To compare exosome separation using the ‘H’method with that using other traditional and commercially available methods, SC-conditioned medium containing exosome-depleted FBS in which SCs were cultured in a 100 mm culture dish was harvested, centrifuged to remove cell debris at 3,000 g for 15 min, and then subjected to each separation method. Isolation was performed using an ultracentrifuge (UC), the ExoQuick-TC ULTRA Kit (company S; System Biosciences), or the Total Exosome Isolation Reagent (T; Thermo Fisher Scientific) according to the manufacturers’ instructions. For exosome separation using a UC, SC-conditioned medium was centrifuged at 10,000 g for 30 min, followed by ultracentrifugation at 100,000 g for 70 min at 4°C (70 Ti rotor; Beckman). The exosomecontaining pellet was washed in coldfiltered PBS and ultracentrifuged again at 100,000 g for 70 min at 4°C. The supernatant was removed and the pellet containing exosome resuspended in filtered PBS.
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was performed using an LM10 (NanoSight) instrument. Samples were diluted with filtered PBS to examine 10~20 vesicles per frame and then gently injected into the laser chamber. Each sample was subjected to a red laser (642 nm) for 1 min three times. The detection threshold was adjusted to 3 to allow detection of nanosized particles. Data were analysed using NTA software (ver. 3.1; Nano Sight). All experiments were conducted at room temperature.
Measurement of cell viability
To examine cell viability in MNs and SCs, we used the LIVE/ DEAD cell staining kit (Abcam) according to the manufacturer’s instructions. Briefly, MNs or SCs were washed three times with PBS, followed by treatment with a mixture of calcein-AM and propidium iodide (PI) in PBS for 30 min. Samples were washed three times with PBS and examined under a confocal microscope (LSM 800; Zeiss). In each experiment, live and dead cells were counted in five random confocal microscopy images.
Exosome induction treatment
For exosome tracking, purified exosomes obtained via the ‘H’method was labeled using the PKH67 fluorescent cell linker kit (Sigma), according to the manufacturer’s instructions with some modifications. Briefly, 15 μL dye solution was diluted with 1 mL solution C and centrifuged at 15,000 g for 10 min to remove aggregated dye. The supernatant was diluted with 1 mL solution C containing EXOSC and incubated for 10 min at room temperature. The mixture was then diluted with 1 mL EXO-FBS and incubated for 10 min, followed by the addition of 10 mL filtered PBS. Then, the solution was centrifuged at 100,000 g for 1 h and the pellets were resuspended with filtered PBS. MNs plated on the coverslip were treated with PKH67- labeled EXOSC and incubated for 24 h and 48 h. The MNs labelled with EXOSC were analyzed by confocal microscopy.
Western blotting analysis
For Western blotting, purified exosomes were mixed with 5% sodium dodecyl sulfate (SDS) sample buffer (T and I), and cells were lysed and homogenized in radioimmunoprecipitation assay (RIPA) buffer (T and I) containing protease inhibitors. Protein concentrations were measured using Bradford assays. Lysed samples were separated onto SDS–polyacrylamide gel electrophoresis (PAGE) gels (12% and 15%) and transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad) for 90 min. The PVDF membrane was blocked with 5% skim milk (BD) in TBST buffer (25 mM Tris, 190 mM NaCl, and 0.05% Tween 20, pH 7.5) for 1~ 2 h at room temperature, followed by treatment with primary antibody at 4°C overnight. After five 20- min washes with TBST, PVDF membranes were incubated with corresponding IgG/horseradish peroxidase (HRP) secondary antibody at a dilution of 1:500 ~ 1:2,000 for 1 h at room temperature, washed, and visualized using the Clarity Western ECL substrate (Bio-Rad). The primary antibodies used were mouse monoclonal anti-CD63 (1:500; Novus), mouse monoclonal anti-TSG101 (1:500; Novus), and mouse monoclonal anti-GAPDH (1:1,000; Novus). After five further 20 min washes with TBST, membranes were incubated with secondary antibody for 1 h at room temperature. Band intensities were measured using ImageJ software (NIH).
Immunofluorescence
All cells were fixed at DIV 7 and 14 with 4% paraformaldehyde (PFA, T and I) at 4°C for 20 min and treated with 0.2% Triton X-100 (Sigma) for 20 min at room temperature, followed by blocking in 4% bovine serum albumin (BSA, Millipore) overnight at 4°C. Fixed samples were incubated overnight with primary antibody diluted in 1% BSA at 4°C, washed three times with 1% BSA and stained with Alexa 488- or 594-conjugated secondary antibodies. The primary antibodies used were antis100 (1:500; Abcam), anti-beta III tubulin (TuJ1; Abcam), and anti-active caspase-3 (1:1,000; Cell Signaling Technology). The secondary antibodies used were goat anti-rabbit IgG H and L (1:1,000; Abcam) and goat antimouse IgG H and L (1:500; Abcam). Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI; Life Technologies) for 15 min. Samples were observed under confocal microscopy. Active caspase-3 fluorescence intensity was examined in confocal images, taken during a single session with the exposure and gain settings kept constant, of samples obtained from one experiment that were processed sideby- side. The number of active caspase-3+ MNs among all MNs was counted in five random fields of view. To quantify SCpositive cells, the number of s100+ cells among all DAPI+ cells was counted in five random fields of view. Three independent experiments were performed. Purified exosomes were mixed with 2% PFA in PBS, plated on a 10 mm coverslip for 1 h, and then washed three times with filtered PBS. Fixed exosomes were blocked with 10% EXO-FBS for 30 min and washed with PBS, followed by treatment with anti-CD63 (1:300) for 1 h at room temperature and washing three times with filtered PBS. Finally, the samples were incubated with goat anti-mouse IgG H and L (1:500; Abcam) for 1 h at room temperature and then mounted using ProLong Gold Antifade Mountant (Thermo Fisher). A solution containing nanovesicles, purified from conditioned medium incubated without cells using a microfluidic system, served as a negative control.
Transmission electron microscopy
Purified exosomes were fixed with 2% glutaraldehyde for overnight at 4°C. The mixture was then diluted 10-fold with PBS for electron microscopic observation. 5 μL of the sample was loaded onto the glow discharged carbon-coated grids (Harrick Plasma, U.S), which were immediately negatively stained using 1% uranyl acetate solution [22]. The grids were examined in a Tecnai 10 transmission electron microscope (FEI, Instrumentation was used in Kangwon Center for systems imaging) operated at 100 kV. Images were collected with a 2k × 2k UltraScan CCD camera (Gatan).
Exosome depletion assays
To assess the depletion of exosomes, cultured SCs were treated with different concentrations of GW4869 at DIV 3 and SCconditioned medium was harvested at DIV 5. SC-conditioned medium under different GW4869 concentrations was separated using a ‘H’method and exosome protein levels were analyzed.
Statistical analyses
All statistical analyses were performed using GraphPad Prism software (GraphPad, Inc.). Analysis of variance (ANOVA) was performed to compare means among three or more groups, and unpaired two-tailed t-tests with Welch’s correction were used to compare means between two groups. Data from a minimum of three independent experiments are presented as means ± standard error of the mean (SEM). Statistical significance was determined at p<0.05.
Separation and identification of SC-derived exosomes
To determine whether primary SCs release exosomes during development, we characterized exosomes separated from SCs. Primary SCs were initially cultured in medium supplemented with 10% FBS for 72 h. Following complement-mediated cytolysis to remove other cell types, such as fibroblasts, conditioned medium was collected, and the culture medium was exchanged for fresh EXO-FBS culture medium for 48 h (Figure 1A). Purified SCs were immunostained with s100, and DAPI was used to stain the nuclei of all cells (Figure 1B). Our results confirmed that 95.2% of DAPI-stained cells were positive for s100 at DIV 5 (Figure 1C). SC-conditioned medium was collected and centrifuged at 3,000 g for 10 min to remove cell debris prior to continuous and noninvasive separation of exosomes from the medium via the ‘H’method [21]. Before using the exosomes separated using the ‘H’method, we examined the purification yield from the ‘H’method and compared it with those obtained using other commercially available exosome isolation methods (UC, company S, and T; Supplementary Figure 1).
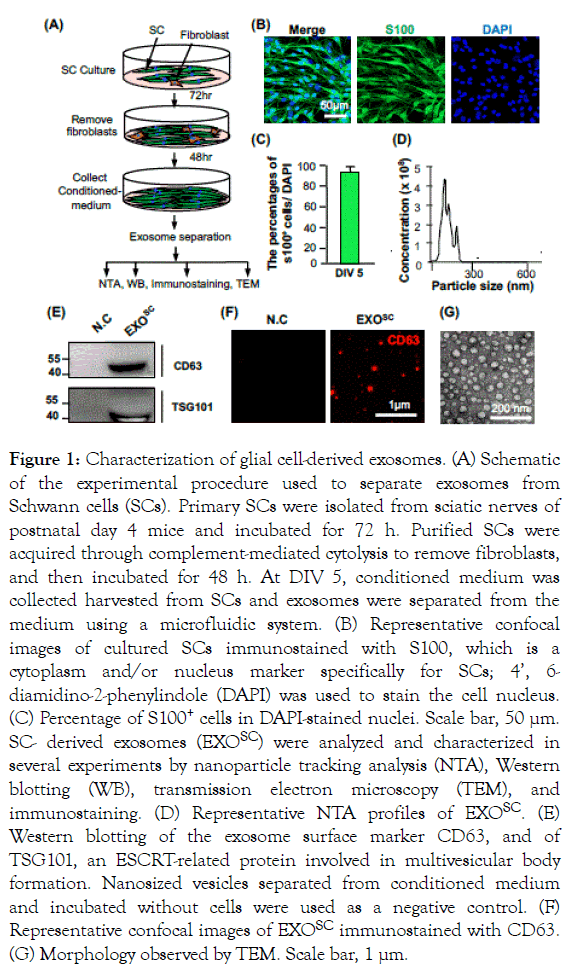
Figure 1. Characterization of glial cell-derived exosomes. (A) Schematic of the experimental procedure used to separate exosomes from Schwann cells (SCs). Primary SCs were isolated from sciatic nerves of postnatal day 4 mice and incubated for 72 h. Purified SCs were acquired through complement-mediated cytolysis to remove fibroblasts, and then incubated for 48 h. At DIV 5, conditioned medium was collected harvested from SCs and exosomes were separated from the medium using a microfluidic system. (B) Representative confocal images of cultured SCs immunostained with S100, which is a cytoplasm and/or nucleus marker specifically for SCs; 4’, 6- diamidino-2-phenylindole (DAPI) was used to stain the cell nucleus. (C) Percentage of S100+ cells in DAPI-stained nuclei. Scale bar, 50 μm. SC- derived exosomes (EXOSC) were analyzed and characterized in several experiments by nanoparticle tracking analysis (NTA), Western blotting (WB), transmission electron microscopy (TEM), and immunostaining. (D) Representative NTA profiles of EXOSC. (E) Western blotting of the exosome surface marker CD63, and of TSG101, an ESCRT-related protein involved in multivesicular body formation. Nanosized vesicles separated from conditioned medium and incubated without cells were used as a negative control. (F) Representative confocal images of EXOSC immunostained with CD63. (G) Morphology observed by TEM. Scale bar, 1 μm.
We confirmed that exosome separation via the ‘H’method resulted in a higher purification yield, and that the level of CD63, a protein marker of exosomes, was 6.6-, 1.4-, and 3.0-fold higher than the yields obtained using a UC, company S, and T, respectively. We then characterized the particles obtained during separation by performing NTA, Western blotting, immunostaining, and TEM. NTA revealed an average vesicle size range expected for these vesicles [23,24]; the total concentration of nanosized particles was 4.2 × 109 particles/mL (Figures 1D and 1E). Western blotting results showed that CD63 and TSG10 were expressed in separated exosomes, and were not detected in the negative control; nanoparticles positive for CD63 were detected in EXOSC. The morphology of EXOSC was confirmed by immunostaining of CD63, which was evident in numerous nanosized particles among the EXOSC. To examine whether exosomes purified from donor cells can be taken up by recipient cells, separated exosomes were labeled with PKH67 (green) and added to neurons. PKH67-positive EXOSC were detected around the nucleus in neurons stained with DAPI (blue), and the labeling efficacy was maintained for 48 h after adding the fluorescent exosomes to the neuronal culture medium (Supplementary Figure 2).
Furthermore, the TEM analysis of EXOSC morphology confirmed that EXOSC were perfectly round in shape (Figure 1G).
MN viability enhanced by EXOSC without SC coculturing
A previous study showed that the MN viability was markedly enhanced by indirect co-culturing of SCs with neurons. To determine whether increased MN viability was mainly mediated by neurotrophic factors or EVs, i.e., exosomes, we cultured MNs, in the presence or absence of EXOSC or SCs, in a Transwell system (Figure 2A). MNs were cultured on a coverslip placed in a Transwell compartment (bottom region) and treated with or without EXOSC. As a control, we indirectly co-cultured MNs and SCs: MNs were plated on a coverslip and placed in the bottom of the Transwell and SCs were seeded on the upper part of the Transwell. The viability of cultured MNs was measured using a calcein-AM (green)/PI (red) staining kit at DIV 7 and 14 (Figure 2B). In the indirect SC–MN co-culture, MN viability was well maintained, with 90.2 ± 1.19% of MNs surviving to DIV 14. In the monoculture without EXOSC, 84.9 ± 2.39% of MNs survived to DIV 7, although viability decreased dramatically to 34.1 ± 5.60% by DIV 14. In contrast, when EXOSC was added to the MN monoculture, MN viability was 90.8 ± 1.56% at DIV 7 and 78.4 ± 6.70% at DIV 14 (Figure 2C). These results suggest that EXOSC markedly enhances MN viability, likely due to the secretion of factors that provide trophic support.
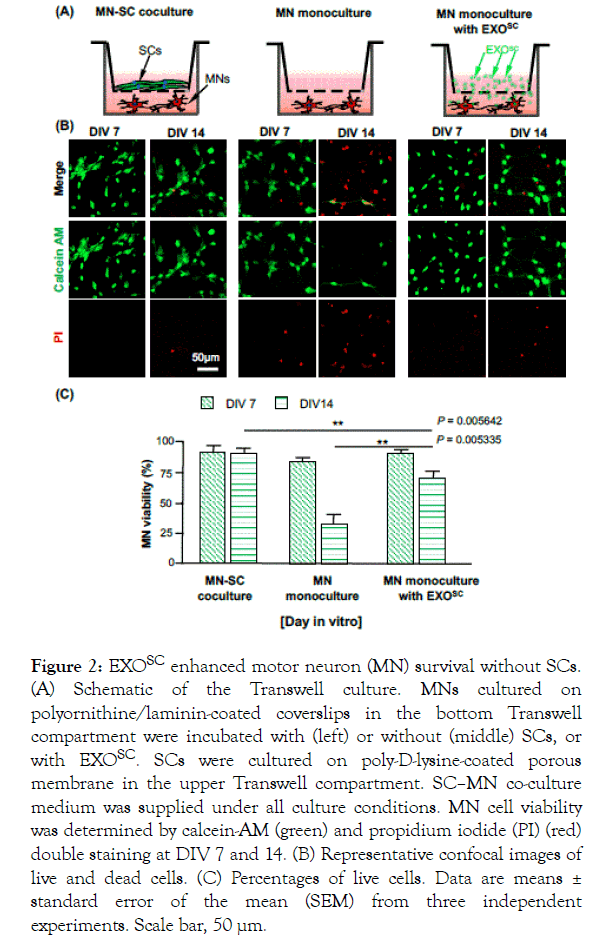
Figure 2. EXOSC enhanced motor neuron (MN) survival without SCs. (A) Schematic of the Transwell culture. MNs cultured on polyornithine/laminin-coated coverslips in the bottom Transwell compartment were incubated with (left) or without (middle) SCs, or with EXOSC. SCs were cultured on poly-D-lysine-coated porous membrane in the upper Transwell compartment. SC–MN co-culture medium was supplied under all culture conditions. MN cell viability was determined by calcein-AM (green) and propidium iodide (PI) (red) double staining at DIV 7 and 14. (B) Representative confocal images of live and dead cells. (C) Percentages of live cells. Data are means ± standard error of the mean (SEM) from three independent experiments. Scale bar, 50 μm.
EXOSC protects MNs from caspase-3 dependent degeneration
A previous study showed that MN death is mediated by caspase-3 activation [17]. To examine whether enhanced MN viability via EXOSC treatment was related to caspase-3 signaling, we observed the number of MNs positive for active caspase-3 at DIV 7 and 14. Consistent with the prominent decrease in MN cell viability from DIV 7 to 14, we observed a large increase in the number of MNs positive for active caspase-3 in the MN monoculture without EXOSC (Figure 3A). Caspase-3 activation at DIV 14 was markedly suppressed by EXOSC treatment; the percentage of caspase-3+ MNs among total MNs exhibited a 2- fold decrease compared with that of MNs without EXOSC (Figure 3B). We also found that treatment with z-DEVD-FMK, a caspase-3 inhibitor, considerably attenuated MN death in the MN monoculture without EXOSC, but had no effect on MN viability following treatment with either EXOSC or SC–MN coculture. These results suggest that EXOSC supports MN survival by preventing the activation of caspase-3-dependent death signalling.
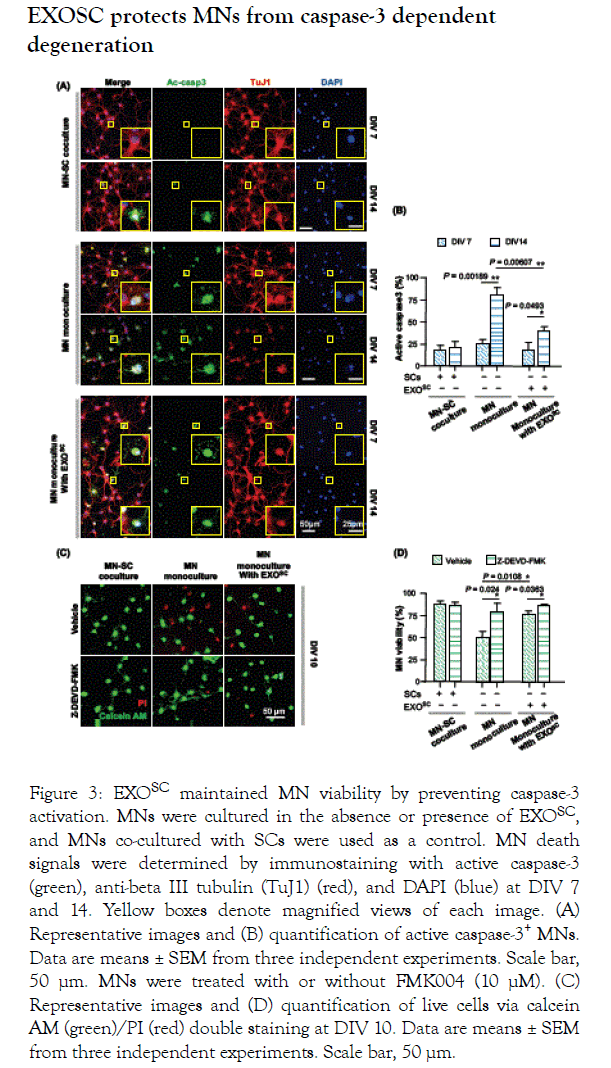
Figure 3. EXOSC maintained MN viability by preventing caspase-3 activation. MNs were cultured in the absence or presence of EXOSC, and MNs co-cultured with SCs were used as a control. MN death signals were determined by immunostaining with active caspase-3 (green), anti-beta III tubulin (TuJ1) (red), and DAPI (blue) at DIV 7 and 14. Yellow boxes denote magnified views of each image. (A) Representative images and (B) quantification of active caspase-3+ MNs. Data are means ± SEM from three independent experiments. Scale bar, 50 μm. MNs were treated with or without FMK004 (10 μM). (C) Representative images and (D) quantification of live cells via calcein AM (green)/PI (red) double staining at DIV 10. Data are means ± SEM from three independent experiments. Scale bar, 50 μm.
Depletion of exosome secretion from SCs prevents MN cell survival
The results described above showed that factors secreted from SCs supported MN survival (SC–MN co-culture; Figure 2B and C), and that the SC secretome includes proteins and, particularly, non-protein components via exosomes; furthermore, EXOSC promotes MN viability (MN monoculture with EXOSC; Figures 2B and 2C). Therefore, we next examined whether the observed increase in MN viability was mediated by EXOSC. To specifically inhibit exosome secretion from cells, SCs were treated with different concentrations of GW4869 (0, 10, 20, and 30 μM) to prevent exosome biogenesis from the multivesicular body.
Nanosized particles were then separated from SC-conditioned medium treated with GW4869 using the ‘H’method. To prevent cell toxicity by GW4869, we analyzed SC viability before observing the depletion of exosomes from SCs treated with GW4869. SC viability was not affected by GW4869 treatment, and there was no significant difference in cell viability among the different GW4869 concentrations (Figures 4A and 4B). CD63 protein expression levels gradually decreased as the GW4869 concentration increased (Figure 4C), and CD63 protein expression at 20 μM GW4869 was about 4.99-fold lower than that without GW4869 (Figure 4D). Furthermore, reduced exosome secretion was induced by GW4869 in a dosedependent manner, and was completely prevented at 20 μM GW4869 (Figures 4C and 4D).
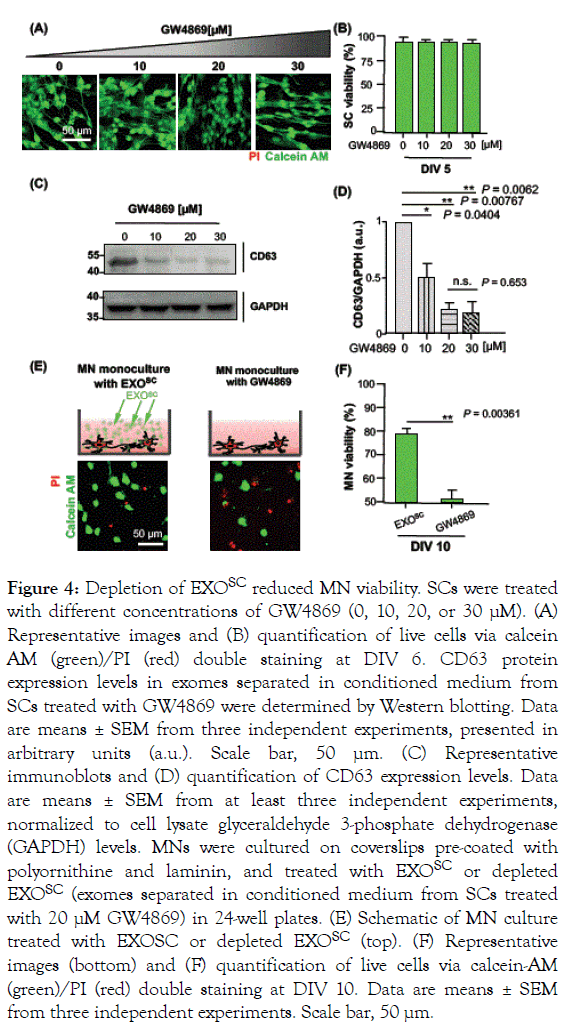
Figure 4. Depletion of EXOSC reduced MN viability. SCs were treated with different concentrations of GW4869 (0, 10, 20, or 30 μM). (A) Representative images and (B) quantification of live cells via calcein AM (green)/PI (red) double staining at DIV 6. CD63 protein expression levels in exomes separated in conditioned medium from SCs treated with GW4869 were determined by Western blotting. Data are means ± SEM from three independent experiments, presented in arbitrary units (a.u.). Scale bar, 50 μm. (C) Representative immunoblots and (D) quantification of CD63 expression levels. Data are means ± SEM from at least three independent experiments, normalized to cell lysate glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. MNs were cultured on coverslips pre-coated with polyornithine and laminin, and treated with EXOSC or depleted EXOSC (exomes separated in conditioned medium from SCs treated with 20 μM GW4869) in 24-well plates. (E) Schematic of MN culture treated with EXOSC or depleted EXOSC (top). (F) Representative images (bottom) and (F) quantification of live cells via calcein-AM (green)/PI (red) double staining at DIV 10. Data are means ± SEM from three independent experiments. Scale bar, 50 μm.
To further confirm that enhanced MN viability was induced by EXOSC, we prevented exosome secretion from SCs using 20 μM GW4869. We observed MN viability at DIV 10 in MNs treated with EXOSC or GW4869, which depleted EXOSC (Figure 4E). MN viability was enhanced following treatment with EXOSC, and reduced under exosome depletion conditions, suggesting that EXOSC provides trophic support to MNs. Importantly, we observed a substantial (1.55-fold) increase in MN survival with EXOSC (EXOSC: 80.3 ± 1.58%, GW4869: 51.6 ± 3.64%) (Figure 4F). Together, these results provide further support for the hypothesis that EXOSC possess neurotrophic factors that can contribute to MN viability.
In this study, we confirmed that SC-secreted exosomes play an essential role in increasing MN cell viability. MN survival was enhanced by treatment of MNs with SC-conditioned medium containing neurotrophic factors, such as BDNF, glial cell linederived neurotrophic factor, neurotophin-3, and nerve growth factor, which promote neuron survival and axon growth [25-28]. Several studies have reported that SCs protect cells from apoptosis and enhance axon regeneration in injured neurons [16,29,30]. Previously, EXOSC derived from human RSC96 SCs showed multiple beneficial effects, including enhanced senescence and cell proliferation, and inhibition of apoptosis in DRG in a cyclic mechanical strain-induced nerve injury model [30]. Axon regeneration in DRG after axotomy was accelerated by treatment with EXOSC [16,29]. Consistent with these findings, our results support the hypothesis that EXOSC mediates MN viability, and perhaps also MN metabolism and other functions.
Exosome-mediated glia-to-neuron signaling has been found to be involved in the regulation of neurotrophic function, and in neuroprotection, under various physiological and pathological conditions [16,31,32]. CNS glia-derived exosomes can reduce the neuronal cell death signaling induced by both oxidative stress and ischemic conditions, by altering the activity of exosomal cargo such as prion protein [31] and apolipoprotein D [32]. Importantly, the large contribution of glia cell-derived exosomes to neuroprotection against neuronal disease has been confirmed through inhibition of exosome cargo, suggesting that exosomes participate in axon–glia communication. In the PNS, because SC-conditioned medium considerably promotes MN cell viability in Transwell co-culture [17], and because we could not determine whether this effect originates from exosomes or neurotrophic factors present in the extracellular space, we favored the hypothesis that SC-induced secretome expression, including of exosomes and neurotrophic factors, has beneficial effects for MN survival and glia–neuron interactions. Cancer cell exosomes similarly control the growth and proliferation of tumor cells through caspase-3-dependent cleavage of Bcl-xL within the exosome [33]. In particular, the caspse-3 signaling pathway appears to be required for exosome uptake by recipient cells, promoting antiapoptotic functions such as cell defense mechanism [33-36]; however, the physiological significance of this pathway in the context of exosomes remains unclear. Consistent with previous studies, we demonstrated that EXOSC support MN survival, in a process mediated by the prevention of activated-caspase-3 signaling. Further experiments are required to determine how exosomes modulate the inhibition of activated-caspase-3 during neuron development.
In the present study, EXOSC significantly improved MN cell viability; this effect was confirmed by depleting the exosome supply. The neuroprotective effect of exosomes on MNs was related to inhibition of the activated-caspase-3 signaling pathway; however, detailed miRNA analysis, particularly of the miRNA content of EXOSC, must be performed to determine the mechanism underlying this process. Nevertheless, these exosome-mediated effects provide new insight into nerve function in the PNS, and point to the potential of exosomes as a therapeutic agent. To elucidate the role of exosomes in neuroprotection, we must examine the fate of exosomes after secretion, to determine whether they are internalized by surrounding neurons or glial cells.
This research was supported by Basic Science Research Program (2018R1A2B6005354 to JWH) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, Future Planning of Korea and National Research Foundation of Korea (NRF-2018M3A9H1023323 to JWH, NRF-2018R1D1A1B07045580to HSJ).
The authors declare no conflict of interests.
Citation: Hyung S, Kim JY, Yu CJ, Jung HS, Hong JW (2019) Neuroprotective Effect of Glial Cell-Derived Exosomes on Neurons. Immunotherapy (Los Angel) 5:156. doi:10.35248/2471-9552.19.5.156
Received: 03-Sep-2019 Accepted: 11-Sep-2019 Published: 18-Sep-2019 , DOI: 10.35248/2471-9552.19.5.156
Copyright: © 2019 Hyung S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests: The authors have declared that no competing interests exist.
Sources of funding : This research was supported by Basic Science Research Program (2018R1A2B6005354 to JWH) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, Future Planning of Korea and National Research Foundation of Korea (NRF-2018M3A9H1023323 to JWH, NRF-2018R1D1A1B07045580to HSJ).